The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila - PubMed (original) (raw)
The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila
T W Burke et al. Genes Dev. 1997.
Abstract
We analyzed the function of the downstream promoter element (DPE), a distinct 7-nucleotide core promoter element that is approximately 30 nucleotides downstream of the transcription start site of many TATA-box-deficient (TATA-less) promoters in Drosophila. There is a strict requirement for spacing between the Inr and DPE motifs, as an increase or decrease of 3 nucleotides in the distance between the Inr and DPE causes a seven- to eightfold reduction in transcription as well as a significant reduction in the binding of purified TFIID. These results suggest a specific and somewhat rigid interaction of TFIID with the Inr and DPE sequences. Photo-cross-linking analysis of purified TFIID with a TATA-less DPE-containing promoter revealed specific cross-linking of dTAFII60 and dTAFII40 to the DPE, with a higher efficiency of cross-linking to dTAFII60 than to dTAFII40. These data, combined with the previously well-characterized interactions between the two TAFs and their homology to histones H4 and H3, suggest that a dTAFII60-dTAFII40 heterotetramer binds to the DPE. Human and Drosophila transcription factors exhibit essentially the same requirements for DPE sequence and for Inr-DPE spacing. In addition, the TATA-less promoter of the human interferon regulatory factor-1 (IRF-1) gene contains a DPE that is important for transcriptional activity both in vitro and in cultured cells. Hence, these studies provide evidence for a direct role of TAFs in basal transcription of TATA-less DPE-containing genes and collectively indicate that the DPE is, in many respects, a downstream counterpart to the TATA box that is present in Drosophila to humans.
Figures
Figure 1
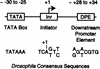
The TATA box, Inr, and DPE are core promoter elements. The consensus sequences and locations of the TATA box, Inr, and DPE motifs are indicated. The TATA box and DPE appear to be functionally redundant, and promoters generally do not contain both elements.
Figure 2
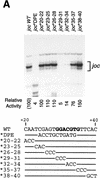
The position of the DPE relative to the Inr is important for transcriptional activity and TFIID binding. (A) In vitro transcription analysis of wild-type and mutant Drosophila joc promoters. A series of mutant joc core promoters (comprising sequences from −3 to +48 relative to the RNA start site) was constructed in which the spacing between the DPE and the Inr was either increased or decreased by 3-nucleotide increments, as depicted. These templates were transcribed in vitro with the Drosophila SK nuclear extract, and the resulting transcripts were subjected to primer extension analysis. The positions of the reverse transcription products from each template are indicated by arrows. Transcriptional activity from the +1 start site for each template is reported as relative to that of the wild-type joc promoter (joc WT). (B) DNase I footprinting analysis of the binding of TFIID to the joc promoter. The mutant versions of the promoter contained either a 3-nucleotide deletion (joc −3) or a 3-nucleotide insertion (joc +3) between the Inr and DPE elements, as in A. The amount of purified Drosophila eTFIID in each reaction is indicated. The positions of the Inr and DPE elements in the mutant promoters are depicted at each side of the autoradiograph.
Figure 3
Analysis of the partial DPE sequence, GA/TCG, in a TATA-less and a TATA-containing promoter. (A) A DPE-related motif in the TATA-less Drosophila Abd-B core promoter is important for transcriptional activity. Wild-type and DPE mutant versions of the Abd-B core promoter (comprising sequences from −17 to +41 relative to the major upstream start site) were constructed, as depicted at the bottom. These templates were transcribed in vitro with the Drosophila SK nuclear extract, and the resulting transcripts were subjected to primer extension analysis. Transcriptional activity is reported as relative to that of the wild-type Abd-B minimal promoter. (B) The DPE-like sequence in the TATA-containing Drosophila hsp70 promoter does not compensate for the loss of the TATA box. Wild-type and mutant versions of the TATA-containing hsp70 core promoter (comprising sequences from −35 to +40 relative to the RNA start site) were constructed, as shown. Transcription reactions were performed with the indicated template DNAs and Drosophila SK nuclear extract. Transcriptional activity is reported as relative to that of the wild-type hsp70 minimal promoter.
Figure 4
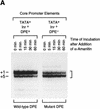
Mutation of the DPE sequence does not affect transcript stability. (A) In vitro transcription reactions were performed with the hb P2–Antp P2 (TATA+Inr+DPE+) and hb P2–Antp P2*DPE (TATA+Inr+DPE−) hybrid minimal promoters (Burke and Kadonaga 1996). After 30-min reaction time, α-amanitin was added (to 4 μg/ml final concentration) to inhibit transcription, and the reaction mixtures were further incubated for the indicated times to determine the stability of the transcripts with the wild-type DPE relative to those with the mutant DPE. The reverse transcription products are indicated by the bracket (right). Note that the preferential use of the +1 start site in the DPE-containing promoter is probably due to the optimal positioning of the DPE to the +1 start site relative to the +5 site, as discussed in Burke and Kadonaga (1996). (B) Graph of data shown in A.
Figure 4
Mutation of the DPE sequence does not affect transcript stability. (A) In vitro transcription reactions were performed with the hb P2–Antp P2 (TATA+Inr+DPE+) and hb P2–Antp P2*DPE (TATA+Inr+DPE−) hybrid minimal promoters (Burke and Kadonaga 1996). After 30-min reaction time, α-amanitin was added (to 4 μg/ml final concentration) to inhibit transcription, and the reaction mixtures were further incubated for the indicated times to determine the stability of the transcripts with the wild-type DPE relative to those with the mutant DPE. The reverse transcription products are indicated by the bracket (right). Note that the preferential use of the +1 start site in the DPE-containing promoter is probably due to the optimal positioning of the DPE to the +1 start site relative to the +5 site, as discussed in Burke and Kadonaga (1996). (B) Graph of data shown in A.
Figure 5
The histone H4-related dTAFII60 is cross-linked to the DPE. (A) dTAFII60 is cross-linked to the joc DPE. Photo-cross-linking experiments were performed with purified TFIID and the joc core promoter. The joc promoter photoprobe was a DNA fragment comprising sequences from −35 to +51 relative to the start site, which contained N3RdUMP in the DPE as well as an adjacent radiolabeled nucleotide (see the +30 probe at the bottom of B). TFIID was incubated with the DNA photoprobe, and the samples were subjected to UV irradiation. The complexes were then digested with nucleases to remove excess DNA, and the resulting radiolabeled proteins were resolved by electrophoresis on a 10% polyacrylamide–SDS gel and detected by autoradiography (left). The migration of each of the polypeptides that comprise TFIID is indicated. Western blot analysis of the cross-linked protein further confirmed that the major radiolabeled band corresponds to dTAFII60 (right). Weaker, yet clearly apparent cross-linking to a species that comigrates with dTAFII40 is also indicated. (B) Sequence-specific photo-cross-linking of the dTAFII60 subunit of TFIID to the wild-type DPE, but not to a mutant DPE. Four different joc promoter photoprobes, which are indicated at the bottom, were each incubated with TFIID and subjected to cross-linking. The positions of N3RdUMP residues (arrows) and radiolabeled nucleotides (stars) are indicated. In the mutant DPE (+30*DPE) probe, the wild-type DPE motif GGACGTG was changed to CTAGCGT. (C) The interaction of dTAFII60 with the DPE photoaffinity probe can be specifically inhibited by competition with a DPE-containing promoter fragment. TFIID was photo-cross-linked to the +30 DPE probe (see B) in the absence or the presence of the indicated amounts (reported as the molar ratio of competitor DNA to probe DNA) of wild-type DPE or mutant DPE promoter DNA fragments. The wild-type DPE competitor DNA fragment was identical to the photoprobe, except that it was not radiolabeled and contained a thymidine residue instead of N3RdUMP. The corresponding mutant DPE competitor DNA contained GGACGTG to CTAGCGT substitutions at the DPE. (D) Photo-cross-linking of TFIID to the DPE in the Antp P2 core promoter. The positions of N3RdUMP residues (arrows) and radiolabeled nucleotides (stars) are indicated.
Figure 5
The histone H4-related dTAFII60 is cross-linked to the DPE. (A) dTAFII60 is cross-linked to the joc DPE. Photo-cross-linking experiments were performed with purified TFIID and the joc core promoter. The joc promoter photoprobe was a DNA fragment comprising sequences from −35 to +51 relative to the start site, which contained N3RdUMP in the DPE as well as an adjacent radiolabeled nucleotide (see the +30 probe at the bottom of B). TFIID was incubated with the DNA photoprobe, and the samples were subjected to UV irradiation. The complexes were then digested with nucleases to remove excess DNA, and the resulting radiolabeled proteins were resolved by electrophoresis on a 10% polyacrylamide–SDS gel and detected by autoradiography (left). The migration of each of the polypeptides that comprise TFIID is indicated. Western blot analysis of the cross-linked protein further confirmed that the major radiolabeled band corresponds to dTAFII60 (right). Weaker, yet clearly apparent cross-linking to a species that comigrates with dTAFII40 is also indicated. (B) Sequence-specific photo-cross-linking of the dTAFII60 subunit of TFIID to the wild-type DPE, but not to a mutant DPE. Four different joc promoter photoprobes, which are indicated at the bottom, were each incubated with TFIID and subjected to cross-linking. The positions of N3RdUMP residues (arrows) and radiolabeled nucleotides (stars) are indicated. In the mutant DPE (+30*DPE) probe, the wild-type DPE motif GGACGTG was changed to CTAGCGT. (C) The interaction of dTAFII60 with the DPE photoaffinity probe can be specifically inhibited by competition with a DPE-containing promoter fragment. TFIID was photo-cross-linked to the +30 DPE probe (see B) in the absence or the presence of the indicated amounts (reported as the molar ratio of competitor DNA to probe DNA) of wild-type DPE or mutant DPE promoter DNA fragments. The wild-type DPE competitor DNA fragment was identical to the photoprobe, except that it was not radiolabeled and contained a thymidine residue instead of N3RdUMP. The corresponding mutant DPE competitor DNA contained GGACGTG to CTAGCGT substitutions at the DPE. (D) Photo-cross-linking of TFIID to the DPE in the Antp P2 core promoter. The positions of N3RdUMP residues (arrows) and radiolabeled nucleotides (stars) are indicated.
Figure 5
The histone H4-related dTAFII60 is cross-linked to the DPE. (A) dTAFII60 is cross-linked to the joc DPE. Photo-cross-linking experiments were performed with purified TFIID and the joc core promoter. The joc promoter photoprobe was a DNA fragment comprising sequences from −35 to +51 relative to the start site, which contained N3RdUMP in the DPE as well as an adjacent radiolabeled nucleotide (see the +30 probe at the bottom of B). TFIID was incubated with the DNA photoprobe, and the samples were subjected to UV irradiation. The complexes were then digested with nucleases to remove excess DNA, and the resulting radiolabeled proteins were resolved by electrophoresis on a 10% polyacrylamide–SDS gel and detected by autoradiography (left). The migration of each of the polypeptides that comprise TFIID is indicated. Western blot analysis of the cross-linked protein further confirmed that the major radiolabeled band corresponds to dTAFII60 (right). Weaker, yet clearly apparent cross-linking to a species that comigrates with dTAFII40 is also indicated. (B) Sequence-specific photo-cross-linking of the dTAFII60 subunit of TFIID to the wild-type DPE, but not to a mutant DPE. Four different joc promoter photoprobes, which are indicated at the bottom, were each incubated with TFIID and subjected to cross-linking. The positions of N3RdUMP residues (arrows) and radiolabeled nucleotides (stars) are indicated. In the mutant DPE (+30*DPE) probe, the wild-type DPE motif GGACGTG was changed to CTAGCGT. (C) The interaction of dTAFII60 with the DPE photoaffinity probe can be specifically inhibited by competition with a DPE-containing promoter fragment. TFIID was photo-cross-linked to the +30 DPE probe (see B) in the absence or the presence of the indicated amounts (reported as the molar ratio of competitor DNA to probe DNA) of wild-type DPE or mutant DPE promoter DNA fragments. The wild-type DPE competitor DNA fragment was identical to the photoprobe, except that it was not radiolabeled and contained a thymidine residue instead of N3RdUMP. The corresponding mutant DPE competitor DNA contained GGACGTG to CTAGCGT substitutions at the DPE. (D) Photo-cross-linking of TFIID to the DPE in the Antp P2 core promoter. The positions of N3RdUMP residues (arrows) and radiolabeled nucleotides (stars) are indicated.
Figure 5
The histone H4-related dTAFII60 is cross-linked to the DPE. (A) dTAFII60 is cross-linked to the joc DPE. Photo-cross-linking experiments were performed with purified TFIID and the joc core promoter. The joc promoter photoprobe was a DNA fragment comprising sequences from −35 to +51 relative to the start site, which contained N3RdUMP in the DPE as well as an adjacent radiolabeled nucleotide (see the +30 probe at the bottom of B). TFIID was incubated with the DNA photoprobe, and the samples were subjected to UV irradiation. The complexes were then digested with nucleases to remove excess DNA, and the resulting radiolabeled proteins were resolved by electrophoresis on a 10% polyacrylamide–SDS gel and detected by autoradiography (left). The migration of each of the polypeptides that comprise TFIID is indicated. Western blot analysis of the cross-linked protein further confirmed that the major radiolabeled band corresponds to dTAFII60 (right). Weaker, yet clearly apparent cross-linking to a species that comigrates with dTAFII40 is also indicated. (B) Sequence-specific photo-cross-linking of the dTAFII60 subunit of TFIID to the wild-type DPE, but not to a mutant DPE. Four different joc promoter photoprobes, which are indicated at the bottom, were each incubated with TFIID and subjected to cross-linking. The positions of N3RdUMP residues (arrows) and radiolabeled nucleotides (stars) are indicated. In the mutant DPE (+30*DPE) probe, the wild-type DPE motif GGACGTG was changed to CTAGCGT. (C) The interaction of dTAFII60 with the DPE photoaffinity probe can be specifically inhibited by competition with a DPE-containing promoter fragment. TFIID was photo-cross-linked to the +30 DPE probe (see B) in the absence or the presence of the indicated amounts (reported as the molar ratio of competitor DNA to probe DNA) of wild-type DPE or mutant DPE promoter DNA fragments. The wild-type DPE competitor DNA fragment was identical to the photoprobe, except that it was not radiolabeled and contained a thymidine residue instead of N3RdUMP. The corresponding mutant DPE competitor DNA contained GGACGTG to CTAGCGT substitutions at the DPE. (D) Photo-cross-linking of TFIID to the DPE in the Antp P2 core promoter. The positions of N3RdUMP residues (arrows) and radiolabeled nucleotides (stars) are indicated.
Figure 6
Human transcription factors can mediate basal transcription via a Drosophila DPE. (A) The conserved DPE consensus sequence is required for basal transcription by HeLa factors. A systematic set of clustered triple point mutant versions of the joc minimal promoter (from −3 to +48 relative to the major RNA start site; Burke and Kadonaga 1996) was subjected to in vitro transcription analysis with a HeLa nuclear extract. The joc promoter sequence from +20 to +40 relative to the transcription start site is shown with the corresponding nucleotide substitutions below. Lines indicate unchanged sequences. The DPE consensus sequence is shown in boldface type. The transcriptional activity is reported relative to that of the wild-type joc minimal (−3 to +48) promoter. (B) The spacing between the Inr and the DPE elements is important for basal transcription by HeLa factors. A series of mutant joc promoters in which the spacing between the Inr and DPE is varied by 3-nucleotide increments, as depicted in Fig. 2A, was subjected to in vitro transcription analysis with a HeLa nuclear extract. The reverse transcription products are indicated by arrows. Transcriptional activity from the +1 start site of each promoter is reported relative to that of the wild-type joc minimal promoter.
Figure 6
Human transcription factors can mediate basal transcription via a Drosophila DPE. (A) The conserved DPE consensus sequence is required for basal transcription by HeLa factors. A systematic set of clustered triple point mutant versions of the joc minimal promoter (from −3 to +48 relative to the major RNA start site; Burke and Kadonaga 1996) was subjected to in vitro transcription analysis with a HeLa nuclear extract. The joc promoter sequence from +20 to +40 relative to the transcription start site is shown with the corresponding nucleotide substitutions below. Lines indicate unchanged sequences. The DPE consensus sequence is shown in boldface type. The transcriptional activity is reported relative to that of the wild-type joc minimal (−3 to +48) promoter. (B) The spacing between the Inr and the DPE elements is important for basal transcription by HeLa factors. A series of mutant joc promoters in which the spacing between the Inr and DPE is varied by 3-nucleotide increments, as depicted in Fig. 2A, was subjected to in vitro transcription analysis with a HeLa nuclear extract. The reverse transcription products are indicated by arrows. Transcriptional activity from the +1 start site of each promoter is reported relative to that of the wild-type joc minimal promoter.
Figure 7
The mammalian IRF-1 gene has a TATA-less, DPE-containing core promoter. (A) The promoters of the IRF-1 genes from mice (m) and humans (h) contain a consensus DPE sequence that is located downstream of the Inr element with spacing that is identical to that seen in many Drosophila DPE-containing promoters. The mammalian Inr and Drosophila DPE consensus sequences are indicated. (B) The hIRF-1 DPE sequence is important for basal transcription in the context of either a minimal core promoter or a larger promoter fragment. The human IRF-1 promoter constructions, which contained the indicated sequences relative to the RNA start site, were transcribed with a HeLa nuclear extract. In the mutant promoter, the DPE sequence was altered from AGACGTG to CTCATGT. The amount of transcription from each mutant promoter is reported as the percentage of transcription from the corresponding wild-type promoter. (C) The Drosophila Antp P2 DPE sequence is important for basal transcription in the context of either a minimal core promoter or a larger promoter fragment. The Antp P2 promoter constructions, which contained the indicated sequences relative to the RNA start site, were transcribed with a Drosophila SK nuclear ex-tract. In the mutant promoter, the DPE sequence was altered from AGACGTG to CTCATGT. The amount of transcription from each mutant promoter is reported as the percentage of transcription from the corresponding wild-type promoter.
Figure 8
The DPE sequence in the human IRF-1 promoter is important for IFN-γ-induced transcription in cultured cells. The hIRF-1 promoter (containing sequences from −1312 to +39 relative to the transcription start site) with a CAT reporter gene was transfected into K562 cells, which were then grown in the presence or the absence of IFN-γ (1500 U/ml), as indicated. The promoterless vector was transfected in parallel as a control. CAT activity is reported relative to that seen with the wild-type promoter in the presence of IFN-γ.
Figure 9
A model for the interaction of TFIID with TATA box-containing vs. TATA-less, DPE-containing promoters. Depicted are two postulated TFIID–promoter interactions, not excluding other types of functionally important interactions between TFIID and core promoters. The lines between TFIID and the Inr indicate interactions between components of TFIID, such as TAFII150 and/or TAFII250, and the Inr.
Similar articles
- Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters.
Burke TW, Kadonaga JT. Burke TW, et al. Genes Dev. 1996 Mar 15;10(6):711-24. doi: 10.1101/gad.10.6.711. Genes Dev. 1996. PMID: 8598298 - The downstream promoter element DPE appears to be as widely used as the TATA box in Drosophila core promoters.
Kutach AK, Kadonaga JT. Kutach AK, et al. Mol Cell Biol. 2000 Jul;20(13):4754-64. doi: 10.1128/MCB.20.13.4754-4764.2000. Mol Cell Biol. 2000. PMID: 10848601 Free PMC article. - The downstream activation sequence of the strict late Herpes Simplex Virus Type 1 U(L)38 promoter interacts with hTAF(II)70, a component of TFIID.
Petroski MD, Devi-Rao GB, Rice MK, Wagner EK. Petroski MD, et al. Virus Genes. 2001 Jun;22(3):299-310. doi: 10.1023/a:1011162106727. Virus Genes. 2001. PMID: 11450948 - The DPE, a core promoter element for transcription by RNA polymerase II.
Kadonaga JT. Kadonaga JT. Exp Mol Med. 2002 Sep 30;34(4):259-64. doi: 10.1038/emm.2002.36. Exp Mol Med. 2002. PMID: 12515390 Review. - The DPE, a conserved downstream core promoter element that is functionally analogous to the TATA box.
Burke TW, Willy PJ, Kutach AK, Butler JE, Kadonaga JT. Burke TW, et al. Cold Spring Harb Symp Quant Biol. 1998;63:75-82. doi: 10.1101/sqb.1998.63.75. Cold Spring Harb Symp Quant Biol. 1998. PMID: 10384272 Review. No abstract available.
Cited by
- Perspectives on the RNA polymerase II core promoter.
Kadonaga JT. Kadonaga JT. Wiley Interdiscip Rev Dev Biol. 2012 Jan-Feb;1(1):40-51. doi: 10.1002/wdev.21. Epub 2011 Dec 6. Wiley Interdiscip Rev Dev Biol. 2012. PMID: 23801666 Free PMC article. Review. - Mapping key functional sites within yeast TFIID.
Leurent C, Sanders SL, Demény MA, Garbett KA, Ruhlmann C, Weil PA, Tora L, Schultz P. Leurent C, et al. EMBO J. 2004 Feb 25;23(4):719-27. doi: 10.1038/sj.emboj.7600111. Epub 2004 Feb 12. EMBO J. 2004. PMID: 14765106 Free PMC article. - Transcriptional regulation of PP2A-A alpha is mediated by multiple factors including AP-2alpha, CREB, ETS-1, and SP-1.
Chen HG, Han WJ, Deng M, Qin J, Yuan D, Liu JP, Xiao L, Gong L, Liang S, Zhang J, Liu Y, Li DW. Chen HG, et al. PLoS One. 2009 Sep 14;4(9):e7019. doi: 10.1371/journal.pone.0007019. PLoS One. 2009. PMID: 19750005 Free PMC article. - Transcriptional and structural impact of TATA-initiation site spacing in mammalian core promoters.
Ponjavic J, Lenhard B, Kai C, Kawai J, Carninci P, Hayashizaki Y, Sandelin A. Ponjavic J, et al. Genome Biol. 2006;7(8):R78. doi: 10.1186/gb-2006-7-8-R78. Epub 2006 Aug 17. Genome Biol. 2006. PMID: 16916456 Free PMC article. - Structural analysis and dimerization potential of the human TAF5 subunit of TFIID.
Bhattacharya S, Takada S, Jacobson RH. Bhattacharya S, et al. Proc Natl Acad Sci U S A. 2007 Jan 23;104(4):1189-94. doi: 10.1073/pnas.0610297104. Epub 2007 Jan 16. Proc Natl Acad Sci U S A. 2007. PMID: 17227857 Free PMC article.
References
- Bartholomew B, Tinker RL, Kassavetis GA, Geiduschek EP. Photochemical cross-linking assay for DNA tracking by replication proteins. Methods Enzymol. 1995;262:476–494. - PubMed
- Biggin MD, Tjian R. Transcription factors that activate the Ultrabithorax promoter in developmentally staged extracts. Cell. 1988;53:699–711. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases