Alu insertion polymorphisms and human evolution: evidence for a larger population size in Africa - PubMed (original) (raw)
Comparative Study
Alu insertion polymorphisms and human evolution: evidence for a larger population size in Africa
M Stoneking et al. Genome Res. 1997 Nov.
Abstract
Alu insertion polymorphisms (polymorphisms consisting of the presence/absence of an Alu element at a particular chromosomal location) offer several advantages over other nuclear DNA polymorphisms for human evolution studies. First, they are typed by rapid, simple, PCR-based assays; second, they are stable polymorphisms-newly inserted Alu elements rarely undergo deletion; third, the presence of an Alu element represents identity by descent-the probability that different Alu elements would independently insert into the exact same chromosomal location is negligible; and fourth, the ancestral state is known with certainty to be the absence of an Alu element. We report here a study of 8 loci in 1500 individuals from 34 worldwide populations. African populations exhibit the most between-population differentiation, and the population tree is rooted in Africa; moreover, the estimated effective time of separation of African versus non-African populations is 137,000 +/- 15,000 years ago, in accordance with other genetic data. However, a principal coordinates analysis indicates that populations from Sahul (Australia and New Guinea) are nearly as close to the hypothetical ancestor as are African populations, suggesting that there was an early expansion of tropical populations of our species. An analysis of heterozygosity versus genetic distance suggests that African populations have had a larger effective population size than non-African populations. Overall, these results support the African origin of modern humans in that an earlier expansion of the ancestors of African populations is indicated.
Figures
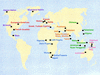
Figure 1
Map of sample localities.
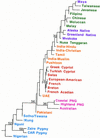
Figure 2
Neighbor-joining tree of population relationships. This tree is rooted where a hypothetical ancestral population—in which the frequency of the Alu element at each of the eight loci is set to 0.0—attaches to the unrooted network. Numbers indicate (in per cent) the fraction of 500 bootstrap replicates that supported a particular grouping.
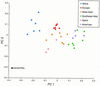
Figure 3
Plot of the first two principal coordinates of the allele frequencies at the eight polymorphic Alu insertion loci. The same relative population relationships were observed when the hypothetical ancestral population (consisting of allele frequencies of zero for the presence of the Alu element at each locus) was removed from the analysis (data not shown), indicating that the inclusion of the ancestral population is not distorting the population relationships.
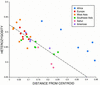
Figure 4
Plot of heterozygosity vs. distance from the centroid. The broken line is the expected relationship predicted by the model of Harpending and Ward (1982), according to the formula _h_i = H(1 − _r_i), where _r_i is the distance from the centroid and _h_i and H are the heterozygosities of population i and the total population, respectively.
Similar articles
- African origin of human-specific polymorphic Alu insertions.
Batzer MA, Stoneking M, Alegria-Hartman M, Bazan H, Kass DH, Shaikh TH, Novick GE, Ioannou PA, Scheer WD, Herrera RJ, et al. Batzer MA, et al. Proc Natl Acad Sci U S A. 1994 Dec 6;91(25):12288-92. doi: 10.1073/pnas.91.25.12288. Proc Natl Acad Sci U S A. 1994. PMID: 7991620 Free PMC article. - The X chromosome Alu insertions as a tool for human population genetics: data from European and African human groups.
Athanasiadis G, Esteban E, Via M, Dugoujon JM, Moschonas N, Chaabani H, Moral P. Athanasiadis G, et al. Eur J Hum Genet. 2007 May;15(5):578-83. doi: 10.1038/sj.ejhg.5201797. Epub 2007 Feb 28. Eur J Hum Genet. 2007. PMID: 17327877 - Genetic structure in African populations: implications for human demographic history.
Lambert CA, Tishkoff SA. Lambert CA, et al. Cold Spring Harb Symp Quant Biol. 2009;74:395-402. doi: 10.1101/sqb.2009.74.053. Epub 2010 May 7. Cold Spring Harb Symp Quant Biol. 2009. PMID: 20453204 Free PMC article. Review. - Evolutionary impact of human Alu repetitive elements.
Jurka J. Jurka J. Curr Opin Genet Dev. 2004 Dec;14(6):603-8. doi: 10.1016/j.gde.2004.08.008. Curr Opin Genet Dev. 2004. PMID: 15531153 Review.
Cited by
- Transposable element-mediated rearrangements are prevalent in human genomes.
Balachandran P, Walawalkar IA, Flores JI, Dayton JN, Audano PA, Beck CR. Balachandran P, et al. Nat Commun. 2022 Nov 19;13(1):7115. doi: 10.1038/s41467-022-34810-8. Nat Commun. 2022. PMID: 36402840 Free PMC article. - Short Tandem Repeats as a High-Resolution Marker for Capturing Recent Orangutan Population Evolution.
Voicu AA, Krützen M, Bilgin Sonay T. Voicu AA, et al. Front Bioinform. 2021 Aug 16;1:695784. doi: 10.3389/fbinf.2021.695784. eCollection 2021. Front Bioinform. 2021. PMID: 36303734 Free PMC article. - An Alu insertion map of the Indian population: identification and analysis in 1021 genomes of the IndiGen project.
Prakrithi P, Singhal K, Sharma D, Jain A, Bhoyar RC, Imran M, Senthilvel V, Divakar MK, Mishra A, Scaria V, Sivasubbu S, Mukerji M. Prakrithi P, et al. NAR Genom Bioinform. 2022 Feb 15;4(1):lqac009. doi: 10.1093/nargab/lqac009. eCollection 2022 Mar. NAR Genom Bioinform. 2022. PMID: 35178516 Free PMC article. - Translational genomics and beyond in bipolar disorder.
Zhang C, Xiao X, Li T, Li M. Zhang C, et al. Mol Psychiatry. 2021 Jan;26(1):186-202. doi: 10.1038/s41380-020-0782-9. Epub 2020 May 18. Mol Psychiatry. 2021. PMID: 32424235 Review. - Identification of a functional human-unique 351-bp Alu insertion polymorphism associated with major depressive disorder in the 1p31.1 GWAS risk loci.
Liu W, Li W, Cai X, Yang Z, Li H, Su X, Song M, Zhou DS, Li X, Zhang C, Shao M, Zhang L, Yang Y, Zhang Y, Zhao J, Chang H, Yao YG, Fang Y, Lv L, Li M, Xiao X. Liu W, et al. Neuropsychopharmacology. 2020 Jun;45(7):1196-1206. doi: 10.1038/s41386-020-0659-2. Epub 2020 Mar 20. Neuropsychopharmacology. 2020. PMID: 32193514 Free PMC article.
References
- Arcot SS, Fontius JJ, Deininger PL, Batzer MA. Identification and analysis of a “young” polymorphic Alu element. Biochem Biophys Acta. 1995a;1263:99–102. - PubMed
- Arcot SS, Wang Z, Weber JL, Deininger PL, Batzer MA. Alu repeats: A source for the genesis of primate microsatellites. Genomics. 1995b;29:136–144. - PubMed
- Arcot SS, Adamson AW, Lamerdin JE, Kanagy B, Deininger PL, Carrano AV, Batzer MA. Alu fossil relics—Distribution and insertion polymorphism. Genome Res. 1996;6:1084–1092. - PubMed
- Armour JAL, Anttinen T, May CA, Vega EE, Sajantila A, Kidd JR, Kidd KK, Bertranpetit J, Paabo S, Jeffreys AJ. Minisatellite diversity supports a recent African origin for modern humans. Nature Genet. 1996;13:154–160. - PubMed
- Batzer MA, Deininger PL. A human-specific subfamily of Alu sequences. Genomics. 1991;9:481–487. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources