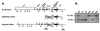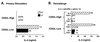An interleukin 4 (IL-4)-independent pathway for CD4+ T cell IL-4 production is revealed in IL-4 receptor-deficient mice - PubMed (original) (raw)
An interleukin 4 (IL-4)-independent pathway for CD4+ T cell IL-4 production is revealed in IL-4 receptor-deficient mice
N Noben-Trauth et al. Proc Natl Acad Sci U S A. 1997.
Abstract
IL-4 receptor alpha chain (IL-4Ralpha)-deficient mice were generated by gene-targeting in BALB/c embryonic stem cells. Mutant mice showed a loss of IL-4 signal transduction and functional activity. The lack of IL-4Ralpha resulted in markedly diminished, but not absent, TH2 responses after infection with the helminthic parasite Nippostrongylus brasiliensis. CD4+, CD62L-high, and CD62L-low T cell populations from uninfected IL-4Ralpha-/- mice were isolated by cell sorting. Upon primary stimulation by T cell receptor cross-linkage, the CD62L-low, but not the CD62L-high, cells secreted considerable amounts of IL-4, which was strikingly enhanced upon 4-day culture with anti-CD3 in the presence or absence of IL-4. CD62L-low cells isolated from IL-4Ralpha-/-, beta2-microglobulin-/- double homozygous mice produced less IL-4 than did either IL-4Ralpha-/- or wild-type mice. These results indicate that an IL-4-independent, beta2-microglobulin-dependent pathway exists through which the CD62L-low CD4+ population has acquired IL-4-producing capacity in vivo, strongly suggesting that these cells are NK T cells.
Figures
Figure 1
Targeting of IL-4Rα. The IL-4Rα locus was disrupted in BALB/c ES cells. (A) The IL-4Rα gene, targeting construct, and locus after homologous recombination are shown. Exons 7–9 (4.5 kb) were replaced by a PGKneo gene orientated antiparallel to the IL-4Rα gene. The targeting vector contained 6.3-kb homology to the genomic sequence. (B) Southern blot analysis of _Eco_RI-digested DNA obtained from offspring of heterozygous matings. The 12.7-kb band indicates the presence of the wild-type allele. The 4.9-kb band reveals the disrupted IL-4Rα locus.
Figure 2
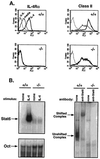
IL-4Rα is not expressed in targeted mice. (A) IL-4Rα expression and induction of IL-4Rα and class II MHC were measured by flow cytometry. Spleen cells from IL-4Rα−/− mice and wild-type littermates were cultured at 1 × 106/ml in cRPMI medium for 40 hr in medium alone or with IL-4 (1000 units/ml). For detection of IL-4Rα, splenocytes were stained with 20 μg/ml rat anti-mouse IL-4Rα (M1) and detected with biotinylated mouse anti-rat IgG2a in the presence of 10 μl normal mouse serum, followed by streptavidin-PE. Class II MHC Iab,d expression was gated on B220+ cells. Dotted line, unstained cells; thin line, medium alone; bold line, with IL-4. (B) STAT6 EMSA. Spleen cells were cultured with IL-4 (10,000 units) for 10 min. Cell lysates were incubated with a double-stranded STAT6-element probe and run on a acrylamide gel. Oct1 sequences were used to control for loading. A polyclonal anti-STAT6 antibody supershifted the complex (Right).
Figure 3
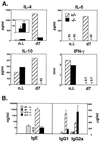
TH2 cytokines are reduced after N. brasiliensis infection. (A) Mesenteric lymph nodes were removed from noninfected controls (n.i.) and mice infected 7 days with N. brasiliensis. Purified CD4+ cells (106/ml) were stimulated for 24 hr with immobilized anti-CD3 and supernatant cytokine content measured by ELISA. (Inset) The graph compares IL-4 production in noninfected (Left) and infected (Right) IL-4Rα−/− mice. (B) Serum IgE, IgG1, and IgG2a were measured at days 0 and 13 after infection. Error bars represent standard deviations (n = 3).
Figure 4
IL-4 is produced by CD62L-low CD4+ cells independent of IL-4 priming. (A) Purified CD4+ T cells were sorted for CD4+, CD62L-high and CD4+, CD62L-low populations. Cells were stimulated (106/ml) for 24 hr with immobilized anti-CD3 and supernatants measured by ELISA. (B) CD4+ T cells were purified and sorted as described and cultured with soluble anti-CD3 with T- depleted splenocytes from BALB/c IL-4−/− mice along with IL-2, anti-IFN-γ, anti-IL-12, and either IL-4 or anti-IL-4 (11B11). After 4 days, cells were washed and rechallenged (106/ml) for 24 hr with immobilized anti-CD3, and supernatants were assayed for IL-4 production.
Figure 5
CD4+, NK1.1+ T cells produce IL-4 in IL-4Rα−/− mice. (A) For measurement of IL-4 from CD4+, NK1.1+ T cells, mice were injected i.v. with anti-CD3. After 90 min spleens were removed and cells cultured for 1 hr. Supernatants were measured for IL-4 by bioassay. Error bars represent standard deviations from three individual mice. (B) CD4 T cells were purified from mesenteric lymph node cells from offspring of IL-4Rα+/− × β2m+/− matings. Cells were sorted for CD4+, CD62L-low populations and stimulated (106/ml) for 24 hr with immobilized anti-CD3 and supernatants measured for IL-4 and IL-2 by ELISA.
Similar articles
- Interleukin-4 receptor alpha-deficient BALB/c mice show an unimpaired T helper 2 polarization in response to Leishmania major infection.
Mohrs M, Holscher C, Brombacher F. Mohrs M, et al. Infect Immun. 2000 Apr;68(4):1773-80. doi: 10.1128/IAI.68.4.1773-1780.2000. Infect Immun. 2000. PMID: 10722563 Free PMC article. - Conventional, naive CD4+ T cells provide an initial source of IL-4 during Th2 differentiation.
Noben-Trauth N, Hu-Li J, Paul WE. Noben-Trauth N, et al. J Immunol. 2000 Oct 1;165(7):3620-5. doi: 10.4049/jimmunol.165.7.3620. J Immunol. 2000. PMID: 11034364 - Deletion of IL-4Ralpha on CD4 T cells renders BALB/c mice resistant to Leishmania major infection.
Radwanska M, Cutler AJ, Hoving JC, Magez S, Holscher C, Bohms A, Arendse B, Kirsch R, Hunig T, Alexander J, Kaye P, Brombacher F. Radwanska M, et al. PLoS Pathog. 2007 May 11;3(5):e68. doi: 10.1371/journal.ppat.0030068. PLoS Pathog. 2007. PMID: 17500591 Free PMC article. - Single cell analysis reveals that IL-4 receptor/Stat6 signaling is not required for the in vivo or in vitro development of CD4+ lymphocytes with a Th2 cytokine profile.
Jankovic D, Kullberg MC, Noben-Trauth N, Caspar P, Paul WE, Sher A. Jankovic D, et al. J Immunol. 2000 Mar 15;164(6):3047-55. doi: 10.4049/jimmunol.164.6.3047. J Immunol. 2000. PMID: 10706693 - Interleukin 4: signalling mechanisms and control of T cell differentiation.
Paul WE. Paul WE. Ciba Found Symp. 1997;204:208-16; discussion 216-9. doi: 10.1002/9780470515280.ch14. Ciba Found Symp. 1997. PMID: 9107423 Review.
Cited by
- Mice Lacking Alternatively Activated (M2) Macrophages Show Impairments in Restorative Sleep after Sleep Loss and in Cold Environment.
Massie A, Boland E, Kapás L, Szentirmai É. Massie A, et al. Sci Rep. 2018 Jun 5;8(1):8625. doi: 10.1038/s41598-018-26758-x. Sci Rep. 2018. PMID: 29872141 Free PMC article. - IL-4 and IL-4 receptor expression is dispensable for the development and function of natural killer T cells.
Sharma A, Berga-Bolanos R, Sultana DA, Sen JM. Sharma A, et al. PLoS One. 2013 Aug 26;8(8):e71872. doi: 10.1371/journal.pone.0071872. eCollection 2013. PLoS One. 2013. PMID: 23990998 Free PMC article. - Neuronal IL-4Rα modulates neuronal apoptosis and cell viability during the acute phases of cerebral ischemia.
Lee HK, Koh S, Lo DC, Marchuk DA. Lee HK, et al. FEBS J. 2018 Aug;285(15):2785-2798. doi: 10.1111/febs.14498. Epub 2018 May 24. FEBS J. 2018. PMID: 29756681 Free PMC article. - Mast cell progenitor trafficking and maturation.
Hallgren J, Gurish MF. Hallgren J, et al. Adv Exp Med Biol. 2011;716:14-28. doi: 10.1007/978-1-4419-9533-9_2. Adv Exp Med Biol. 2011. PMID: 21713649 Free PMC article. Review. - Priming for T helper type 2 differentiation by interleukin 2-mediated induction of interleukin 4 receptor alpha-chain expression.
Liao W, Schones DE, Oh J, Cui Y, Cui K, Roh TY, Zhao K, Leonard WJ. Liao W, et al. Nat Immunol. 2008 Nov;9(11):1288-96. doi: 10.1038/ni.1656. Epub 2008 Sep 28. Nat Immunol. 2008. PMID: 18820682 Free PMC article.
References
- Paul W E. Blood. 1991;77:1859–1870. - PubMed
- Keegan A D, Nelms K, Wang L M, Pierce J H, Paul W E. Immunol Today. 1994;15:423–432. - PubMed
- Beckmann M P, Cosman D, Fanslow W, Maliszewski C R, Lyman S D. In: Interleukins: Molecular Biology and Immunology. Kishimoto T, editor. Basel: Karger; 1992. pp. 107–134. - PubMed
- Kondo M, Takeshita T, Ishii N, Nakamura M, Watanabe S, Arai K-I, Sugamura K. Science. 1993;262:1874–1877. - PubMed
- Russell S M, Keegan A D, Harada N, Nakamura Y, Noguchi M, Leland P, Friedmann M C, Miyajima A, Puri R K, Paul W E, Leonard W J. Science. 1993;262:1880–1883. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials