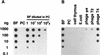Localization of the modified base J in telomeric VSG gene expression sites of Trypanosoma brucei - PubMed (original) (raw)
Localization of the modified base J in telomeric VSG gene expression sites of Trypanosoma brucei
F van Leeuwen et al. Genes Dev. 1997.
Abstract
African trypanosomes such as Trypanosoma brucei undergo antigenic variation in the bloodstream of their mammalian hosts by regularly changing the variant surface glycoprotein (VSG) gene expressed. The transcribed VSG gene is invariably located in a telomeric expression site. There are multiple expression sites and one way to change the VSG gene expressed is by activating a new site and inactivating the previously active one. The mechanisms that control expression site switching are unknown, but have been suggested to involve epigenetic regulation. We have found previously that VSG genes in silent (but not active) expression sites contain modified restriction endonuclease cleavage sites, and we have presented circumstantial evidence indicating that this is attributable to the presence of a novel modified base beta-D-glucosyl-hydroxymethyluracil, or J. To directly test this, we have generated antisera that specifically recognize J-containing DNA and have used these to determine the precise location of this modified thymine in the telomeric VSG expression sites. By anti J-DNA immunoprecipitations, we found that J is present in telomeric VSG genes in silenced expression sites and not in actively transcribed telomeric VSG genes. J was absent from inactive chromosome-internal VSG genes. DNA modification was also found at the boundaries of expression sites. In the long 50-bp repeat arrays upstream of the promoter and in the telomeric repeat arrays downstream of the VSG gene, J was found both in silent and active expression sites. This suggests that silencing results in a gradient of modification spreading from repetitive DNA flanks into the neighboring expression site sequences. In this paper, we discuss the possible role of J in silencing of expression sites.
Figures
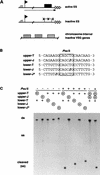
Figure 1
The modified base J prevents cleavage by _Pvu_II. (A) Partial cleavage by restriction enzymes _Pst_I, _Pvu_II, and other enzymes (see text) suggested that silenced expression sites acquire a DNA modification (X) in and around the inactive telomeric VSG gene (box). These blocked restriction sites are not found in transcribed expression sites (broken line with arrowhead) or in silent chromosome-internal VSG genes. Telomeric repeats are indicated by triangles, the expression site promoter by a flag, and the imperfect tandem 50-bp repeats by a hatched box. (B) The effect of substitution of T by J on digestion of duplex DNA by _Pvu_II restriction endonuclease; 20-mer oligonucleotides with a central _Pvu_II restriction site (box) were used as substrates. If present, J replaced T in the _Pvu_II site in either the upper or the lower strand, or replaced T two positions downstream of the _Pvu_II site in the lower strand (lower-J*). (C) End-labeled oligomers (circled plus signs) were annealed to their nonlabeled complementary strands (+) to form duplex oligomers (ds), which were incubated without (−) or with (+) _Pvu_II enzyme. Substrate and products were separated by native 20% polyacrylamide gel electrophoresis. Cleavage at 37°C resulted in short fragments that melted to single-stranded molecules at this temperature (cleaved ss). The last two lanes show a mixing control of duplexes with and without J. Incubation with _Pvu_II (+) resulted in a noncleaved and a cleaved product.
Figure 2
Detection of J-containing DNA on filters with polyclonal anti-J antisera. Dot-blots with dilution series of denatured DNA were incubated with polyclonal antiserum 539αJ and bound antibodies were detected with sheep α-rabbit-HRP in combination with enhanced chemiluminescence. (A) The sensitivity of detection was determined with a dilution series of BF trypanosome DNA (0.2 mole% J) and PC DNA (no J), and BF DNA diluted in PC DNA. Less than 0.0002 mole% J could be detected. Hybridization with a telomere repeat probe showed that equal amounts of DNA were present in each dilution series (data not shown). (B) Specificity of the antisera was tested with DNA samples with various DNA modifications: calf thymus (5-methylC), E. coli (6-methylA, 4-methylC, 5-methylC), phage φe (HOMeU), phage T2 (HOMeC, α-gluc- and β-gluc-α-gluc-HOMeC), phage T4 (α- and β-gluc-HOMeC). Pre-immune sera gave no signal at all (data not shown).
Figure 3
Specific immunoprecipitation of J-containing double-stranded DNA depends on the length of the fragments and the density of modification. (A) DNA fragments of 118, 426, and 943 bp were generated by PCR-amplification with one antisense primer containing one J residue and three different sense primers without J. (B) The 118-bp fragment was used to generate a partial ligation ladder, resulting in fragments of different size with a constant J-density (one J per 118 bp, or no J). End-labeled fragments bound by the J-specific antisera were captured by protein-A beads, separated by 1.5% agarose gel electrophoresis, and blotted onto Hybond-N. 100% of the immunoprecipitated DNA (ip) and 10% of the supernatant (sup) was loaded. (C) Quantitation of the immunoprecipitated fraction as a percentage of the total input (average of three experiments with standard deviation).
Figure 4
Localization of J at the telomeric end of VSG expression sites by anti-J immunoprecipitation. (A) Restriction maps of VSG genes 121, 221, and 1.1, expressed in BF clones 121a, 221a, and r5-1.1 respectively. These genes are present in all clones analyzed here, including the PC clone, which was used as a negative control. In addition to the ELC of VSG 121, three chromosome-internal basic-copy (BC) 121 genes are present in all clones. In clone r5-1.1, the 221 gene has moved from its telomeric position in the expression site to a chromosome-internal position where the _Pvu_II site is no longer modified (see text). (Solid boxes) Coding sequence; (open triangles) telomeric repeat arrays, of which the length varies per clone and per telomere. The VSG gene-specific probes are indicated underneath the coding sequences. (B) _Bgl_I; (C) _Cla_I; (D) _Dra_I; (E) _Eco_RI; (H) _Hin_dIII; (N) _Nco_I. (B) Immunoprecipitation of VSG genes alone. DNA of the clones indicated at the top was digested with restriction enzymes shown on the right of each panel. Modified DNA fragments bound by the anti-J antibodies (ip) and 10% of the supernatant (sup) were analyzed by Southern blot hybridization. The VSG gene on the left of each panel indicates the probe used (probe fragments are shown in A). Note that the one telomeric copy and three basic copies of VSG gene 121 cannot be discriminated in the _Bgl_I–_Nco_I digest used for this VSG gene. (C) Analysis of modification of the three chromosome-internal BCs and the telomeric ELC of VSG gene 121 by αJ-IP of _Hin_dIII restriction fragments. A size marker is indicated on the right. (D) Analysis of the telomeric repeat arrays associated with expression sites. The fragments were detected by hybridization with probes specific for the VSG genes still linked to the telomeric tracts. The restriction digests, the VSG genes probed for, and a size marker are shown on the left of each panel. As a control, PC DNA was mixed with BF 221a DNA and probed for 1.1 to show that nonmodified PC telomeres do not specifically coimmunoprecipitate with modified BF DNA. Note that immunoprecipitation of VSG genes linked to telomeric repeats was consistently more efficient than that of VSG genes alone. Therefore, the smear upstream of the 121 telomere (sup), which is enriched on immunoprecipitation, is most likely attributable to cross-hybridization of the 121 probe to other VSG genes associated with telomeric tracts.
Figure 5
Detection of J in the 50-bp repeats upstream of inactive and active expression sites. Cell lines with a HYG gene in an active (ES2) or inactive (ES2-R1) 221 expression site were used for anti-J immunoprecipitation of sequences in and around the expression site promoter (flag). (N) _Nco_I; (C) _Cla_I; (P) _Hpa_I; (X) _Xba_I. The 5′ part of the HYG gene (line underneath the map) was used as a probe to specifically detect the tagged expression site sequences. The fragments analyzed are indicated on the left of each panel and include from top to bottom HYG gene alone, HYG gene linked to expression site promoter sequences, and HYG gene linked to the 50-bp repeat array. DNA of PC cells with a HYG gene downstream of the expression site promoter (PCES2) was mixed with wild-type BF 221a DNA (BF) as a negative control for nonspecific coimmunoprecipitation of nonmodified DNA. The solid box indicates the _HYG_-coding sequence, stippled boxes RNA-processing signals, and the striped box 50-bp repeats.
Figure 6
The modified base J in and around telomeric VSG expression sites. Schematic representation of the distribution of J in active and inactive VSG expression sites determined by αJ immunoprecipitation of sonicated DNA combined with dot-blot hybridizations (expression site adapted from Revelard et al. 1990). ES probes are described in Materials and Methods. ESAGs were studied as the cumulative signal of all copies in the genome using sonicated DNA. Telomeric repeats, 50-bp repeats, 70-bp repeats, VSG genes, and promoter regions were also studied in individual expression sites using restriction digests. % IP shows a quantitation of the IP efficiency (immunoprecipitated fraction of the input) of expression site sequences using sonicated DNA (average with standard deviation of two independent clones 221a and 221aR12). The numbers correspond to the expression site sequences shown above. IP of silent VSG genes in expression sites varied from 1 (±0.1) to 16.2 (±3.4) depending on the VSG gene studied. The absence of J in actively transcribed VSG genes and 70-bp repeats strongly suggests that J is also absent from active ESAGs, but this has not been tested directly.
Similar articles
- beta-D-glucosyl-hydroxymethyluracil, a novel base in African trypanosomes and other Kinetoplastida.
Borst P, van Leeuwen F. Borst P, et al. Mol Biochem Parasitol. 1997 Dec 1;90(1):1-8. doi: 10.1016/s0166-6851(97)00170-9. Mol Biochem Parasitol. 1997. PMID: 9497027 Review. - Tandemly repeated DNA is a target for the partial replacement of thymine by beta-D-glucosyl-hydroxymethyluracil in Trypanosoma brucei.
van Leeuwen F, Kieft R, Cross M, Borst P. van Leeuwen F, et al. Mol Biochem Parasitol. 2000 Jul;109(2):133-45. doi: 10.1016/s0166-6851(00)00247-4. Mol Biochem Parasitol. 2000. PMID: 10960172 - Biosynthesis and function of the modified DNA base beta-D-glucosyl-hydroxymethyluracil in Trypanosoma brucei.
van Leeuwen F, Kieft R, Cross M, Borst P. van Leeuwen F, et al. Mol Cell Biol. 1998 Oct;18(10):5643-51. doi: 10.1128/MCB.18.10.5643. Mol Cell Biol. 1998. PMID: 9742081 Free PMC article. - A ribosomal DNA promoter replacing the promoter of a telomeric VSG gene expression site can be efficiently switched on and off in T. brucei.
Rudenko G, Blundell PA, Dirks-Mulder A, Kieft R, Borst P. Rudenko G, et al. Cell. 1995 Nov 17;83(4):547-53. doi: 10.1016/0092-8674(95)90094-2. Cell. 1995. PMID: 7585957 - Control of VSG gene expression sites.
Borst P, Ulbert S. Borst P, et al. Mol Biochem Parasitol. 2001 Apr 25;114(1):17-27. doi: 10.1016/s0166-6851(01)00243-2. Mol Biochem Parasitol. 2001. PMID: 11356510 Review.
Cited by
- Dynamic colocalization of 2 simultaneously active VSG expression sites within a single expression-site body in Trypanosoma brucei.
Budzak J, Kerry LE, Aristodemou A, Hall BS, Witmer K, Kushwaha M, Davies C, Povelones ML, McDonald JR, Sur A, Myler PJ, Rudenko G. Budzak J, et al. Proc Natl Acad Sci U S A. 2019 Aug 13;116(33):16561-16570. doi: 10.1073/pnas.1905552116. Epub 2019 Jul 29. Proc Natl Acad Sci U S A. 2019. PMID: 31358644 Free PMC article. - Genome-wide distribution of 5-hydroxymethyluracil and chromatin accessibility in the Breviolum minutum genome.
Marinov GK, Chen X, Swaffer MP, Xiang T, Grossman AR, Greenleaf WJ. Marinov GK, et al. bioRxiv [Preprint]. 2023 Sep 22:2023.09.18.558303. doi: 10.1101/2023.09.18.558303. bioRxiv. 2023. PMID: 37781619 Free PMC article. Updated. Preprint. - Genetic Interaction Between Site-Specific Epigenetic Marks and Roles of H4v in Transcription Termination in Trypanosoma brucei.
Kim HS. Kim HS. Front Cell Dev Biol. 2021 Oct 14;9:744878. doi: 10.3389/fcell.2021.744878. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34722526 Free PMC article. - The epigenome of Trypanosoma brucei: a regulatory interface to an unconventional transcriptional machine.
Maree JP, Patterton HG. Maree JP, et al. Biochim Biophys Acta. 2014 Sep;1839(9):743-50. doi: 10.1016/j.bbagrm.2014.05.028. Epub 2014 Jun 3. Biochim Biophys Acta. 2014. PMID: 24942804 Free PMC article. Review. - Intrinsic DNA curvature in trypanosomes.
Smircich P, El-Sayed NM, Garat B. Smircich P, et al. BMC Res Notes. 2017 Nov 9;10(1):585. doi: 10.1186/s13104-017-2908-y. BMC Res Notes. 2017. PMID: 29121981 Free PMC article.
References
- Bernards A, Van der Ploeg LH, Frasch AC, Borst P, Boothroyd JC, Coleman S, Cross GA. Activation of trypanosome surface glycoprotein genes involves a duplication-transposition leading to an altered 3′ end. Cell. 1981;27:497–505. - PubMed
- Bernards A, Michels PAM, Lincke CR, Borst P. Growth of chromosome ends in multiplying trypanosomes. Nature. 1983;303:592–597. - PubMed
- Bernards A, de Lange T, Michels PA, Liu AY, Huisman MJ, Borst P. Two modes of activation of a single surface antigen gene of Trypanosoma brucei. Cell. 1984a;36:163–170. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources