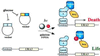A yeast genetic system for selecting small molecule inhibitors of protein-protein interactions in nanodroplets - PubMed (original) (raw)
A yeast genetic system for selecting small molecule inhibitors of protein-protein interactions in nanodroplets
J Huang et al. Proc Natl Acad Sci U S A. 1997.
Abstract
Cellular processes are mediated by complex networks of molecular interactions. Dissection of their role most commonly is achieved by using genetic mutations that alter, for example, protein-protein interactions. Small molecules that accomplish the same result would provide a powerful complement to the genetic approach, but it generally is believed that such molecules are rare. There are several natural products, however, that illustrate the feasibility of this approach. Split-pool synthesis now provides a simple mechanical means to prepare vast numbers of complex, even natural product-like, molecules individually attached to cell-sized polymer beads. Here, we describe a genetic system compatible with split-pool synthesis that allows the detection of cell-permeable, small molecule inhibitors of protein-protein interactions in 100- to 200-nl cell culture droplets, prepared by a recently described technique that arrays large numbers of such droplets. These "nanodroplets" contain defined media, cells, and one or more beads containing approximately 100 pmol of a photoreleasable small molecule and a controlled number of cells. The engineered Saccharomyces cerevisiae cells used in this study express two interacting proteins after induction with galactose whose interaction results in cell death in the presence of 5-fluoroorotic acid (inducible reverse two-hybrid assay). Disruption of the interaction by a small molecule allows growth, and the small molecule can be introduced into the system hours before induction of the toxic interaction. We demonstrate that the interaction between the activin receptor R1 and the immunophilin protein FKBP12 can be disrupted by the small molecule FK506 at nanomolar concentrations in nanodroplets. This system should provide a general method for selecting cell-permeable ligands that can be used to study the relevance of protein-protein interactions in living cells or organisms.
Figures
Figure 1
General schematic of the inducible (small molecule) reverse two-hybrid system designed to detect small molecule inhibitors of protein–protein interactions. The expression of the interacting proteins is controlled by the GAL1 promoter, which is repressed in Glc. After Gal induction, the two interacting protein partners are synthesized, and their association in turn induces the synthesis of a toxic gene product, leading to death, unless an inhibitor of the protein–protein interaction is present in the cell. Only cells with this disruption should be selected.
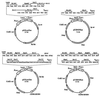
Figure 2
(A) Reporter constructs used for homologous recombination. The LexAop_4–_SPO13
tata
–URA3 reporter construct contains four LexA operators upstream of a SPO13 promoter–URA3 fusion reporter gene. Nucleotide numbering is relative to the translation start codon ATG, where A is +1. Red numbering represents nucleotides from the URA3 gene, and green represents those from the SPO13 gene. (B) Schematic representation of reporter yeast strains.
Figure 3
Vectors for inducible two-hybrid protein expression. Expression of the two interacting proteins is under the control of the GAL1 promoter. Expression vectors contain antibiotic markers for selection. Both centromeric and 2-μ vectors were designed.
Figure 4
FK506 specifically interferes with the interaction between FKBP12 and the activin type I receptor R1 as detected in the small molecule reverse two-hybrid system. Sc* refers to synthetic complete (Sc) medium without any sugar; gal/raf, 2% galactose plus 1% raffinose; and glc, 2% glucose. (A) Phenotypes of R1C–FKBP12 interaction and its dissociation by FK506. Yeast transformants with both vectors appear within 24–36 h on Sc-H-W plates. Colonies were picked and patched on various selective plates for reported gene assays. Each pairwise transformation was analyzed by six independent transformants, three of which are shown here. (B) FK506-mediated 5-FOA resistance is specific to R1C–FKBP12 transformants. (C) Detection of the FK506 effect in liquid media using arrayed nanodroplets (≈200-nl vol) (36). The droplets containing yeast, medium, and FK506 beads were formed in polydimethylsiloxane plastic molded to contain wells 40 μm in depth, 1 mm in diameter, and 250 μm apart from each other.
Similar articles
- Small molecule-dependent genetic selection in stochastic nanodroplets as a means of detecting protein-ligand interactions on a large scale.
Borchardt A, Liberles SD, Biggar SR, Crabtree GR, Schreiber SL. Borchardt A, et al. Chem Biol. 1997 Dec;4(12):961-8. doi: 10.1016/s1074-5521(97)90304-5. Chem Biol. 1997. PMID: 9427663 - TGF-beta-signaling with small molecule FKBP12 antagonists that bind myristoylated FKBP12-TGF-beta type I receptor fusion proteins.
Stockwell BR, Schreiber SL. Stockwell BR, et al. Chem Biol. 1998 Jul;5(7):385-95. doi: 10.1016/s1074-5521(98)90072-2. Chem Biol. 1998. PMID: 9662508 - A three-hybrid system for detecting small ligand-protein receptor interactions.
Licitra EJ, Liu JO. Licitra EJ, et al. Proc Natl Acad Sci U S A. 1996 Nov 12;93(23):12817-21. doi: 10.1073/pnas.93.23.12817. Proc Natl Acad Sci U S A. 1996. PMID: 8917502 Free PMC article. - Mechanism of action of rapamycin: new insights into the regulation of G1-phase progression in eukaryotic cells.
Wiederrecht GJ, Sabers CJ, Brunn GJ, Martin MM, Dumont FJ, Abraham RT. Wiederrecht GJ, et al. Prog Cell Cycle Res. 1995;1:53-71. doi: 10.1007/978-1-4615-1809-9_5. Prog Cell Cycle Res. 1995. PMID: 9552353 Review. - Use of the yeast two-hybrid system for identifying the cascade of protein interactions resulting in apoptotic cell death.
Bemis LT, Geske FJ, Strange R. Bemis LT, et al. Methods Cell Biol. 1995;46:139-51. doi: 10.1016/s0091-679x(08)61928-7. Methods Cell Biol. 1995. PMID: 7609652 Review.
Cited by
- Current Experimental Methods for Characterizing Protein-Protein Interactions.
Zhou M, Li Q, Wang R. Zhou M, et al. ChemMedChem. 2016 Apr 19;11(8):738-56. doi: 10.1002/cmdc.201500495. Epub 2016 Feb 11. ChemMedChem. 2016. PMID: 26864455 Free PMC article. Review. - Resolving the network of cell signaling pathways using the evolving yeast two-hybrid system.
Ratushny V, Golemis E. Ratushny V, et al. Biotechniques. 2008 Apr;44(5):655-62. doi: 10.2144/000112797. Biotechniques. 2008. PMID: 18474041 Free PMC article. Review. - The yeast two-hybrid system and its pharmaceutical significance.
Topcu Z, Borden KL. Topcu Z, et al. Pharm Res. 2000 Sep;17(9):1049-55. doi: 10.1023/a:1026493310144. Pharm Res. 2000. PMID: 11087035 Review. - A yeast two-hybrid system for the screening and characterization of small-molecule inhibitors of protein-protein interactions identifies a novel putative Mdm2-binding site in p53.
Wong JH, Alfatah M, Sin MF, Sim HM, Verma CS, Lane DP, Arumugam P. Wong JH, et al. BMC Biol. 2017 Nov 9;15(1):108. doi: 10.1186/s12915-017-0446-7. BMC Biol. 2017. PMID: 29121928 Free PMC article. - On the binding affinity of macromolecular interactions: daring to ask why proteins interact.
Kastritis PL, Bonvin AM. Kastritis PL, et al. J R Soc Interface. 2012 Dec 12;10(79):20120835. doi: 10.1098/rsif.2012.0835. Print 2013 Feb. J R Soc Interface. 2012. PMID: 23235262 Free PMC article. Review.
References
- Yamamoto K R. Annu Rev Genet. 1985;19:209–252. - PubMed
- Schreiber S L. Science. 1991;251:283–287. - PubMed
- Schreiber S L. Chem Eng News. 1992;26:22–32.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials