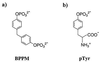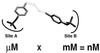Identification of a second aryl phosphate-binding site in protein-tyrosine phosphatase 1B: a paradigm for inhibitor design - PubMed (original) (raw)
Identification of a second aryl phosphate-binding site in protein-tyrosine phosphatase 1B: a paradigm for inhibitor design
Y A Puius et al. Proc Natl Acad Sci U S A. 1997.
Abstract
The structure of the catalytically inactive mutant (C215S) of the human protein-tyrosine phosphatase 1B (PTP1B) has been solved to high resolution in two complexes. In the first, crystals were grown in the presence of bis-(para-phosphophenyl) methane (BPPM), a synthetic high-affinity low-molecular weight nonpeptidic substrate (Km = 16 microM), and the structure was refined to an R-factor of 18. 2% at 1.9 A resolution. In the second, crystals were grown in a saturating concentration of phosphotyrosine (pTyr), and the structure was refined to an R-factor of 18.1% at 1.85 A. Difference Fourier maps showed that BPPM binds PTP1B in two mutually exclusive modes, one in which it occupies the canonical pTyr-binding site (the active site), and another in which a phosphophenyl moiety interacts with a set of residues not previously observed to bind aryl phosphates. The identification of a second pTyr molecule at the same site in the PTP1B/C215S-pTyr complex confirms that these residues constitute a low-affinity noncatalytic aryl phosphate-binding site. Identification of a second aryl phosphate binding site adjacent to the active site provides a paradigm for the design of tight-binding, highly specific PTP1B inhibitors that can span both the active site and the adjacent noncatalytic site. This design can be achieved by tethering together two small ligands that are individually targeted to the active site and the proximal noncatalytic site.
Figures
Figure 1
Structures of PTP1B substrates BPPM (a) and pTyr (b).
Figure 2
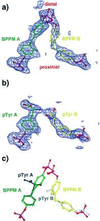
(a and b) Simulated annealing omit maps showing unbiased electron density for bound BPPM (a) and phoshphotyrosine (b) molecules. The density shown is an _F_o − _F_c map contoured at 2.0 σ, with the refined models superimposed. Molecules bound to the canonical pTyr binding site are colored green, and ligands bound to the second site are colored yellow. (c) Superposition of BPPM and pTyr ligands. pTyr A is drawn in black, and pTyr B is gray. [Diagrams were generated with program
o
(16)].
Figure 3
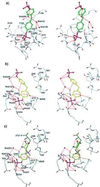
Stereo representations of the binding modes of BPPM A (a), BPPM B (b), and pTyr B (c). Contacts represented by dashed lines are distances less than 3.6 Å, except for certain interactions with aromatic rings. Interactions between the amide nitrogens of residues 216–221 and the phosphate groups of ligand A are too numerous to represent. [Diagrams were generated with the program
o
(16)].
Figure 4
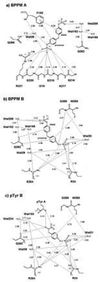
Schematic representations of the interactions between PTP1B/C215S and BPPM A (a), BPPM B (b), and pTyr B (c). A distance cutoff of 3.6 Å was used, except for certain interactions with aromatic rings.
Figure 5
A strategy for creating selective and high-affinity PTP1B inhibitors. Based on the principle of additivity of free energy of binding, high-affinity ligands can be designed by linking two functional groups (each with modest affinity to the target protein) identified experimentally. The added specificity arises from the fact that the tethered ligand has to bind both sites simultaneously.
Similar articles
- Structure of protein tyrosine phosphatase 1B in complex with inhibitors bearing two phosphotyrosine mimetics.
Jia Z, Ye Q, Dinaut AN, Wang Q, Waddleton D, Payette P, Ramachandran C, Kennedy B, Hum G, Taylor SD. Jia Z, et al. J Med Chem. 2001 Dec 20;44(26):4584-94. doi: 10.1021/jm010266w. J Med Chem. 2001. PMID: 11741477 - The structure of apo protein-tyrosine phosphatase 1B C215S mutant: more than just an S --> O change.
Scapin G, Patel S, Patel V, Kennedy B, Asante-Appiah E. Scapin G, et al. Protein Sci. 2001 Aug;10(8):1596-605. doi: 10.1110/ps.11001. Protein Sci. 2001. PMID: 11468356 Free PMC article. - Small molecule interactions with protein-tyrosine phosphatase PTP1B and their use in inhibitor design.
Burke TR Jr, Ye B, Yan X, Wang S, Jia Z, Chen L, Zhang ZY, Barford D. Burke TR Jr, et al. Biochemistry. 1996 Dec 17;35(50):15989-96. doi: 10.1021/bi961256d. Biochemistry. 1996. PMID: 8973169 - Peptidomimetic competitive inhibitors of protein tyrosine phosphatases.
Shen K, Qi L, Stiff L. Shen K, et al. Curr Pharm Des. 2010;16(28):3101-17. doi: 10.2174/138161210793292537. Curr Pharm Des. 2010. PMID: 20687872 Review. - Bidentate inhibitors of protein tyrosine phosphatases.
Low JL, Chai CL, Yao SQ. Low JL, et al. Antioxid Redox Signal. 2014 May 10;20(14):2225-50. doi: 10.1089/ars.2013.5710. Epub 2014 Jan 8. Antioxid Redox Signal. 2014. PMID: 24206395 Review.
Cited by
- Phosphotyrosine isosteres: past, present and future.
Cerulli RA, Kritzer JA. Cerulli RA, et al. Org Biomol Chem. 2020 Jan 28;18(4):583-605. doi: 10.1039/c9ob01998g. Epub 2019 Nov 28. Org Biomol Chem. 2020. PMID: 31777907 Free PMC article. Review. - Advanced Bioinformatics Tools in the Pharmacokinetic Profiles of Natural and Synthetic Compounds with Anti-Diabetic Activity.
Udrea AM, Gradisteanu Pircalabioru G, Boboc AA, Mares C, Dinache A, Mernea M, Avram S. Udrea AM, et al. Biomolecules. 2021 Nov 14;11(11):1692. doi: 10.3390/biom11111692. Biomolecules. 2021. PMID: 34827690 Free PMC article. Review. - Assessment of Antidiabetic Activity of the Shikonin by Allosteric Inhibition of Protein-Tyrosine Phosphatase 1B (PTP1B) Using State of Art: An In Silico and In Vitro Tactics.
Saeed M, Shoaib A, Tasleem M, Alabdallah NM, Alam MJ, Asmar ZE, Jamal QMS, Bardakci F, Alqahtani SS, Ansari IA, Badraoui R. Saeed M, et al. Molecules. 2021 Jun 30;26(13):3996. doi: 10.3390/molecules26133996. Molecules. 2021. PMID: 34208908 Free PMC article. - Protein tyrosine phosphatases--from housekeeping enzymes to master regulators of signal transduction.
Tonks NK. Tonks NK. FEBS J. 2013 Jan;280(2):346-78. doi: 10.1111/febs.12077. Epub 2013 Jan 17. FEBS J. 2013. PMID: 23176256 Free PMC article. Review. - Protein tyrosine phosphatases: structure, function, and implication in human disease.
Tautz L, Critton DA, Grotegut S. Tautz L, et al. Methods Mol Biol. 2013;1053:179-221. doi: 10.1007/978-1-62703-562-0_13. Methods Mol Biol. 2013. PMID: 23860656 Free PMC article. Review.
References
- Hunter T. Cell. 1995;80:225–236. - PubMed
- Tonks N K, Neel B G. Cell. 1996;87:365–368. - PubMed
- Tonks N K, Diltz C D, Fischer E H. J Biol Chem. 1988;263:6731–6737. - PubMed
- Zhang Z-Y, Maclean D, McNamara D J, Dobrusin E M, Sawyer T K, Dixon J E. Biochemistry. 1994;33:2285–2290. - PubMed
- Zhang Z-Y. Curr Top Cell Regul. 1997;35:21–68. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM50121/GM/NIGMS NIH HHS/United States
- T32 GM07288/GM/NIGMS NIH HHS/United States
- R01 CA069202/CA/NCI NIH HHS/United States
- T32 GM007288/GM/NIGMS NIH HHS/United States
- CA69202/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous