Use of differential display analysis to assess the effect of human cytomegalovirus infection on the accumulation of cellular RNAs: induction of interferon-responsive RNAs - PubMed (original) (raw)
Use of differential display analysis to assess the effect of human cytomegalovirus infection on the accumulation of cellular RNAs: induction of interferon-responsive RNAs
H Zhu et al. Proc Natl Acad Sci U S A. 1997.
Abstract
We used differential display analysis to identify mRNAs that accumulate to enhanced levels in human cytomegalovirus-infected cells as compared with mock-infected cells. RNAs were compared at 8 hr after infection of primary human fibroblasts. Fifty-seven partial cDNA clones were isolated, representing about 26 differentially expressed mRNAs. Eleven of the mRNAs were virus-coded, and 15 were of cellular origin. Six of the partial cDNA sequences have not been reported previously. All of the cellular mRNAs identified in the screen are induced by interferon alpha. The induction in virus-infected cells, however, does not involve the action of interferon or other small signaling molecules. Neutralizing antibodies that block virus infection also block the induction. These RNAs accumulate after infection with virus that has been inactivated by treatment with UV light, indicating that the inducer is present in virions. We conclude that human cytomegalovirus induces interferon-responsive mRNAs.
Figures
Figure 1
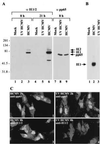
Characterization of UV HCMV. (A) Western blot showing that UV irradiation of virions blocks expression of the HCMV IE1 and IE2 RNAs, but has no effect on the delivery of a virion protein to cells. HF cells were mock-infected or infected, and extracts were prepared 8 or 21 hr later. Lanes 1–6 were reacted with antibody that binds to IE1 and IE2, whereas lanes 7–9 were reacted with antibody to pp65. The molecular weights of marker proteins are indicated on the left. (B) Northern blot showing that IE1 RNA is detected at 8 hr after infection of HF cells with HCMV but not after infection with UV HCMV. (C) Immunofluorescent localization of pp65 and IE1/2 within infected cells. HF cells were infected with HCMV or UV HCMV for 2 or 8 hr, reacted with antibody to pp65 or IE1/IE2 followed by a fluorescein-labeled secondary antibody (bright nuclear signal), and counterstained with ethidium homodimer-1 (dim whole cell signal).
Figure 5
Kinetic analysis of cig RNA accumulation. HF cells were mock-infected (M) or treated with the inducers identified to the right (HCMV, HSV-1, and IFN-α), RNA was prepared at various times after treatment (indicated above lanes), and analyzed by Northern blot by using the probes indicated on the left (cig1, cig49, HCMV IE1, and HSV-1 icp47).
Figure 2
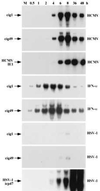
Differential expression of RNAs in HF cells assayed by Northern blot. (A) RNA was prepared from mock-infected (M), HCMV-infected cells (C), or UV HCMV-infected cells (UV) and assayed by using cloned cDNA segments. The different clones (cigs) are identified above the panels. (B) RNA was prepared from mock-infected (M), HCMV-infected (C), or HCMV-infected cells that were treated with cycloheximide (CX) and assayed as in A. (C) RNA was prepared from mock-infected cells (M), HCMV-infected cells (C), or cells treated with interferon α (I) and assayed by using probes corresponding to the cigs or the mxA gene. (D) RNA was prepared as in A and assayed with probes corresponding to interferon-inducible genes (mxA, isg15K, and IFN-β) or control genes that are not induced by interferon (p53, p21, cPLA2, and actin).
Figure 3
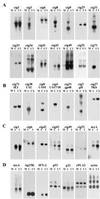
The HCMV particle mediates the induction of differentially expressed HF RNAs. (A) Requirements for induction monitored by Northern blot assay. The relative amounts of three cellular RNAs (cig1, cig6, and cig49) were monitored in mock-infected cells (mock), HCMV AD169-infected cells (HCMV), cells treated with medium from which virions were removed by filtration (inf. med.), mock-infected cells treated with cycloheximide (CX), HCMV AD169-infected cells treated with cycloheximide (CX+HCMV), cells infected with purified HCMV AD169 particles (virions), cells infected with purified noninfectious enveloped particles from HCMV AD169 (NIEPs), adenovirus-infected cells (Ad _dl_309), herpes simplex type 1-infected cells (HSV-1), HCMV Towne-infected cells, HCMV Toledo-infected cells, interferon α-treated cells (IFN-α), and cells treated with interferon that was added to medium and passed through the filter type used to exclude virus (fil. IFN-α). (B) Northern blot assay showing that antibody that neutralizes HCMV (antibody C) blocks the induction of cig RNA accumulation, whereas antibodies that neutralize interferon α or β (antibody Iα, Iβ) block the induction of cig RNAs by interferon α or β (inducer Iα, Iβ) but have no effect on the induction of cig RNAs by HCMV (C). RNA prepared from mock-infected control cells is designated M. The cellular cytosolic phospholipase A2 RNA was assayed as a loading control (control, cPLA2).
Figure 4
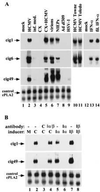
Requirements for the induction of cig RNA accumulation. (A) An intact HCMV particle is required. Purified virions were treated with a mixture of Triton X-100 and deoxycholate (T/C) and separated by centrifugation into supernatant (S) and pellet (P) fractions. Northern blot assays show the effect of detergent treatment on the induction of two cig RNAs (cig1 and cig49) by virions (HCMV) or interferon α (IFN-α). (B) The induction of cig RNAs does not involve the release of mediators stored within infected HF cells. At 8 hr after treatment, RNA was prepared from mock-infected cells (lane 1), HCMV-infected cells (lane 2), or a 9:1 mixture of mock and infected cells (lanes 3 and 4). The two mixed cultures differed in the time of mixing. In lane 3, cells were mixed at 1 hr after infection; in lane 4, RNAs were prepared and mixed from 8 hr mock- and HCMV-infected cells. RNAs were analyzed by Northern blot by using cellular (cig1, cig6, cig49, and cPLA2) and viral (IE1) probes.
Similar articles
- Altered cellular mRNA levels in human cytomegalovirus-infected fibroblasts: viral block to the accumulation of antiviral mRNAs.
Browne EP, Wing B, Coleman D, Shenk T. Browne EP, et al. J Virol. 2001 Dec;75(24):12319-30. doi: 10.1128/JVI.75.24.12319-12330.2001. J Virol. 2001. PMID: 11711622 Free PMC article. - Human cytomegalovirus latent infection alters the expression of cellular and viral microRNA.
Fu M, Gao Y, Zhou Q, Zhang Q, Peng Y, Tian K, Wang J, Zheng X. Fu M, et al. Gene. 2014 Feb 25;536(2):272-8. doi: 10.1016/j.gene.2013.12.012. Epub 2013 Dec 18. Gene. 2014. PMID: 24361963 - Rhesus cytomegalovirus particles prevent activation of interferon regulatory factor 3.
DeFilippis V, Früh K. DeFilippis V, et al. J Virol. 2005 May;79(10):6419-31. doi: 10.1128/JVI.79.10.6419-6431.2005. J Virol. 2005. PMID: 15858025 Free PMC article. - Human cytomegalovirus: a review of developments between 1970 and 1976. Part II. Experimental developments.
Michelson-Fiske S. Michelson-Fiske S. Biomedicine. 1977 Apr;26(2):86-97. Biomedicine. 1977. PMID: 194635 Review. - Alterations in TLRs as new molecular markers of congenital infections with Human cytomegalovirus?
Wujcicka W, Wilczyński J, Nowakowska D. Wujcicka W, et al. Pathog Dis. 2014 Feb;70(1):3-16. doi: 10.1111/2049-632X.12083. Epub 2013 Sep 10. Pathog Dis. 2014. PMID: 23929630 Review.
Cited by
- Unexpected regulatory functions of cyprinid Viperin on inflammation and metabolism.
Chaumont L, Jouneau L, Huetz F, van Muilekom DR, Peruzzi M, Raffy C, Le Hir J, Minke J, Boudinot P, Collet B. Chaumont L, et al. BMC Genomics. 2024 Jun 29;25(1):650. doi: 10.1186/s12864-024-10566-x. BMC Genomics. 2024. PMID: 38951796 Free PMC article. - Plasma proteome perturbation for CMV DNAemia in kidney transplantation.
Sigdel TK, Boada P, Kerwin M, Rashmi P, Gjertson D, Rossetti M, Sur S, Munar D, Cimino J, Ahn R, Pickering H, Sen S, Parmar R, Fatou B, Steen H, Schaenman J, Bunnapradist S, Reed EF, Sarwal MM; CMV Systems Immunobiology Group. Sigdel TK, et al. PLoS One. 2023 May 19;18(5):e0285870. doi: 10.1371/journal.pone.0285870. eCollection 2023. PLoS One. 2023. PMID: 37205661 Free PMC article. - Radical-SAM dependent nucleotide dehydratase (SAND), rectification of the names of an ancient iron-sulfur enzyme using NC-IUBMB recommendations.
Ji Y, Wei L, Da A, Stark H, Hagedoorn PL, Ciofi-Baffoni S, Cowley SA, Louro RO, Todorovic S, Mroginski MA, Nicolet Y, Roessler MM, Le Brun NE, Piccioli M, James WS, Hagen WR, Ebrahimi KH. Ji Y, et al. Front Mol Biosci. 2022 Oct 21;9:1032220. doi: 10.3389/fmolb.2022.1032220. eCollection 2022. Front Mol Biosci. 2022. PMID: 36387278 Free PMC article. No abstract available. - High Glucose Induces in HK2 Kidney Cells an IFN-Dependent ZIKV Antiviral Status Fueled by Viperin.
Reslan A, Haddad JG, Desprès P, Bascands JL, Gadea G. Reslan A, et al. Biomedicines. 2022 Jul 1;10(7):1577. doi: 10.3390/biomedicines10071577. Biomedicines. 2022. PMID: 35884880 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases