RanGTP targets p97 to RanBP2, a filamentous protein localized at the cytoplasmic periphery of the nuclear pore complex - PubMed (original) (raw)
RanGTP targets p97 to RanBP2, a filamentous protein localized at the cytoplasmic periphery of the nuclear pore complex
C Delphin et al. Mol Biol Cell. 1997 Dec.
Free PMC article
Abstract
RanBP2, a protein containing FG repeat motifs and four binding sites for the guanosine triphosphatase Ran, is localized at the cytoplasmic periphery of the nuclear pore complex (NPC) and is believed to play a critical role in nuclear protein import. We purified RanBP2 from rat liver nuclear envelopes and examined its structural and biochemical properties. Electron microscopy showed that RanBP2 forms a flexible filamentous molecule with a length of approximately 36 nm, suggesting that it comprises a major portion of the cytoplasmic fibrils implicated in initial binding of import substrates to the NPC. Using in vitro assays, we characterized the ability of RanBP2 to bind p97, a cytosolic factor implicated in the association of the nuclear localization signal receptor with the NPC. We found that RanGTP promotes the binding of p97 to RanBP2, whereas it inhibits the binding of p97 to other FG repeat nucleoporins. These data suggest that RanGTP acts to specifically target p97 to RanBP2, where p97 may support the binding of an nuclear localization signal receptor/substrate complex to RanBP2 in an early step of nuclear import.
Figures
Figure 1
Purification of RanBP2 from rat liver NE. Proteins obtained during different steps of the purification were analyzed by SDS-PAGE on a 5–20% gradient gel followed by staining with Coomassie blue. (A) Lane 1, molecular weight standards (molecular weights indicated to the left of lane 1); lane 2, total NE; lane 3, salt-washed NE; lane 4, solubilized NE proteins; lane 5, proteins eluted from Ran affinity beads; lane 6, purified RanBP2. Proteins in lanes 2–4 were derived from 1 OD260 NE, and proteins in lanes 5–6 were derived from 20 OD260 NE. (B) Purified RanBP2 (from 20 OD260 NE) was analyzed by silver staining after SDS-PAGE as in panel A
Figure 2
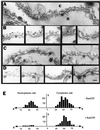
Electron microscopy of negatively stained RanBP2. Purified RanBP2 was adsorbed to glow-discharged carbon-coated grids and was negatively stained with uranyl acetate. Panels A and C are electron micrographs of two different grid preparations that show predominantly an extended (panel A) or folded (panel B) conformation of RanBP2. Arrows indicate examples of partially folded conformations of RanBP2 found in both preparations. Panel C is a gallery of folded RanBP2 molecules selected from the same preparation as panel B. Bar, 100 nm.
Figure 3
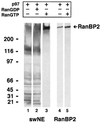
p97 binds selectively to RanBP2 in the presence of RanGTP. A blot overlay approach was used to analyze the binding of p97 to proteins of salt-washed NE (swNE, 2 OD260, lanes 1–3), or to purified RanBP2 (10 ng, lanes 4–5). Protein samples were separated by SDS-PAGE on a 5–15% gradient gel, electrophoretically transferred to nitrocellulose, and the nitrocellulose membrane was renatured. Nitrocellulose strips were incubated with GSTp97, RanGDP, and RanGTP as indicated. After cross-linking, bound p97 was detected using anti-GST antibodies.
Figure 4
RanGTP selectively targets p97 to the cytoplasmic side of the NPC. GST-p97 was incubated with isolated rat liver NE in the absence (A and B) or presence (C and D) of RanGTP. Bound p97 was visualized by labeling with goat anti-GST antibodies followed by rabbit anti-goat antibodies coupled with 5 nm gold. Samples were then processed for thin section electron microscopy. (E) Histograms showing the distance of gold particles from the midplane of the NPC for the binding of p97 in the absence (top) or presence (bottom) of RanGTP. X axis: distance of gold particles from the midplane; Y axis: number of gold particles counted; c, cytoplasm; n, nucleoplasm.
Figure 5
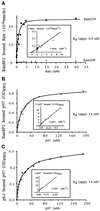
Characterization of the interactions of Ran and p97 with nucleoporins RanBP2 and p62. (A) Titration of Ran binding to RanBP2. Increasing concentrations of Ran loaded with either [32P]GTP or [32P]GDP were incubated with RanBP2 adsorbed to microtiter wells. After washing, the radioactivity was recovered from the wells with SDS and counted in a liquid scintillation counter, and the amount of Ran bound to RanBP2 was calculated from the [32P]GTP-specific activity. (B and C) Titration of p97 binding to RanBP2 and p62, respectively. Increasing concentration of His-p97 were incubated with RanBP2 (B) or p62 (C) and adsorbed to microtiter wells. The 6xHis-p97 that remained after washing was then cross-linked to the adsorbed protein and detected with antibodies (see MATERIALS AND METHODS). The Lineweaver-Burk plots used to determine the apparent binding constants (Kdapp.) are shown in the insets.
Figure 6
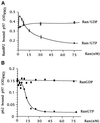
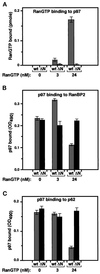
RanGTP modulates the interaction of p97 with RanBP2 and p62. (A) Effect of Ran on the binding of p97 to RanBP2. RanBP2-coated microtiter wells were incubated with 10 nM His-p97 and with an increasing amount of RanGTP or RanGDP. After incubation and washing, proteins were cross-linked, and the amount of bound p97 was quantified using antibodies (see MATERIALS AND METHODS). (B) Effect of Ran on the binding of p97 to p62. The analysis was carried out as in panel A, except that p62-coated microtiter wells were used.
Figure 7
The ability of RanGTP to modulate the binding of p97 to RanBP2 and p62 requires the RanGTP/p97 interaction. (A) 3 nM or 24 nM [32P]GTP-Ran was incubated with wt p97 or ΔN p97 bound to microtiter wells. After binding and washing, the radioactivity was recovered from the plate by elution with SDS and was counted in a liquid scintillation counter. The amount of bound RanGTP was calculated from the [32P]RanGTP-specific activity. (B) The binding of wt p97 and ΔN p97 to RanBP2 adsorbed to microtiter wells was analyzed in the absence of Ran, or in the presence of 3 nM or 24 nM RanGTP, and the bound p97 was quantified as in Figure 4. (C) The binding of wt p97 and ΔN p97 to p62 adsorbed to microtiter wells was analyzed as in panel B.
Similar articles
- RanBP1 stabilizes the interaction of Ran with p97 nuclear protein import.
Chi NC, Adam EJ, Visser GD, Adam SA. Chi NC, et al. J Cell Biol. 1996 Nov;135(3):559-69. doi: 10.1083/jcb.135.3.559. J Cell Biol. 1996. PMID: 8909533 Free PMC article. - Localization of the Ran-GTP binding protein RanBP2 at the cytoplasmic side of the nuclear pore complex.
Wilken N, Senécal JL, Scheer U, Dabauvalle MC. Wilken N, et al. Eur J Cell Biol. 1995 Nov;68(3):211-9. Eur J Cell Biol. 1995. PMID: 8603673 - A role for RanBP1 in the release of CRM1 from the nuclear pore complex in a terminal step of nuclear export.
Kehlenbach RH, Dickmanns A, Kehlenbach A, Guan T, Gerace L. Kehlenbach RH, et al. J Cell Biol. 1999 May 17;145(4):645-57. doi: 10.1083/jcb.145.4.645. J Cell Biol. 1999. PMID: 10330396 Free PMC article. - Nuclear import and export pathways.
Moroianu J. Moroianu J. J Cell Biochem. 1999;Suppl 32-33:76-83. doi: 10.1002/(sici)1097-4644(1999)75:32+<76::aid-jcb10>3.3.co;2-h. J Cell Biochem. 1999. PMID: 10629106 Review. - Molecular dissection of the nuclear pore complex.
Panté N, Aebi U. Panté N, et al. Crit Rev Biochem Mol Biol. 1996 Apr;31(2):153-99. doi: 10.3109/10409239609106583. Crit Rev Biochem Mol Biol. 1996. PMID: 8740526 Review.
Cited by
- Gradient of increasing affinity of importin beta for nucleoporins along the pathway of nuclear import.
Ben-Efraim I, Gerace L. Ben-Efraim I, et al. J Cell Biol. 2001 Jan 22;152(2):411-7. doi: 10.1083/jcb.152.2.411. J Cell Biol. 2001. PMID: 11266456 Free PMC article. - A link between the synthesis of nucleoporins and the biogenesis of the nuclear envelope.
Marelli M, Lusk CP, Chan H, Aitchison JD, Wozniak RW. Marelli M, et al. J Cell Biol. 2001 May 14;153(4):709-24. doi: 10.1083/jcb.153.4.709. J Cell Biol. 2001. PMID: 11352933 Free PMC article. - Distinct RanBP1 nuclear export and cargo dissociation mechanisms between fungi and animals.
Li Y, Zhou J, Min S, Zhang Y, Zhang Y, Zhou Q, Shen X, Jia D, Han J, Sun Q. Li Y, et al. Elife. 2019 Apr 25;8:e41331. doi: 10.7554/eLife.41331. Elife. 2019. PMID: 31021318 Free PMC article. - Herpes simplex virus replication: roles of viral proteins and nucleoporins in capsid-nucleus attachment.
Copeland AM, Newcomb WW, Brown JC. Copeland AM, et al. J Virol. 2009 Feb;83(4):1660-8. doi: 10.1128/JVI.01139-08. Epub 2008 Dec 10. J Virol. 2009. PMID: 19073727 Free PMC article.
References
- Bischoff FR, Ponstingl H. Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature. 1991a;354:80–82. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous