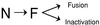Influenza hemagglutinin is spring-loaded by a metastable native conformation - PubMed (original) (raw)
Influenza hemagglutinin is spring-loaded by a metastable native conformation
C M Carr et al. Proc Natl Acad Sci U S A. 1997.
Abstract
Enveloped viruses enter cells by protein-mediated membrane fusion. For influenza virus, membrane fusion is regulated by the conformational state of the hemagglutinin (HA) protein, which switches from a native (nonfusogenic) structure to a fusion-active (fusogenic) conformation when exposed to the acidic environment of the cellular endosome. Here we demonstrate that destabilization of HA at neutral pH, with either heat or the denaturant urea, triggers a conformational change that is biochemically indistinguishable from the change triggered by low pH. In each case, the conformational change is coincident with induction of membrane-fusion activity, providing strong evidence that the fusogenic structure is formed. These results indicate that the native structure of HA is trapped in a metastable state and that the fusogenic conformation is released by destabilization of native structure. This strategy may be shared by other enveloped viruses, including those that enter the cell at neutral pH, and could have implications for understanding the membrane-fusion step of HIV infection.
Figures
Figure 1
Model for the spring-loaded mechanism for membrane fusion (20, 21). Envelope of an influenza virus (bottom membrane) is juxtaposed to the target membrane of the cellular endosome (Top). In the native conformation (Left) an HA trimer facilitates viral attachment via the HA1 subunits (orange). In response to acidic pH (Right), the HA1 subunits dissociate, the loop regions (yellow) become helical and extend the HA2 coiled coil (green), and the fusion-peptide regions (blue) insert into the target membrane. Membrane fusion per se appears to require only the HA2 subunit (2). Figure by Jodi M. Harris (adapted from ref. 80).
Figure 5
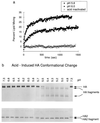
Acid-induced membrane-fusion activation and the HA conformational change. (a) At 37°C, acid-induced activity is detected at pH 6.0 (solid circles), with optimal activity at pH 5.8 (solid squares) and no activity from acid-inactivated virus (open circles). (b) The conformational change detected by immunoblot analysis was induced by acid below pH 6.0, at 37°C.
Figure 6
Heat-induced membrane-fusion activation and the HA conformational change. (a) At pH 7.3, heat-induced activity is detected at 58°C (solid circles), with optimal activity at 60°C (solid squares) and no activity from heat-inactivated virus (open circles). (b) The conformational change detected by immunoblot analysis was induced by heat above 58°C at pH 7.3.
Figure 7
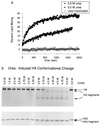
Denaturant-induced membrane-fusion activation and the HA conformational change. (a) At pH 7.3 and 37°C, denaturant-induced activity is detected at 3.0 M urea (solid circles), with optimal activity at 3.5 M urea (solid squares) and no activity from urea-inactivated virus (open circles). (b) The conformational change detected by immunoblot analysis was induced by urea above 3.5M at 37°C, pH 7.3.
Figure 2
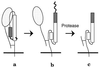
Schematic of the protease assay for the conformational change of HA. In one monomer of the HA trimer, the HA1 polypeptide (stippled ball and string) is connected to the HA2 polypeptide by a disulfide bond. The HA2 polypeptide spans the viral membrane (Bottom) once and includes the fusion peptide region. (a) Native HA is resistant to proteolysis. (b) The conformational change: HA1 dissociates from the top of HA2, and the fusion peptide is propelled to the top of the molecule. (c) Proteolysis of the fusogenic conformation: some of HA1 and the fusion peptide of HA2 are proteolytically degraded, leaving the majority of HA2 disulfide-bonded to a fragment of HA1. As a result, both the HA1 and the HA2 polypeptides are smaller, and this difference can be detected by SDS/PAGE and immunoblot analysis.
Figure 3
Conditions that induce the HA conformational change. The protease assay is used to monitor the conformation of HA on virus exposed to experimental conditions (indicated above the gel lanes). (a) Viral polypeptides are separated by SDS/PAGE and detected by Coomassie blue staining of viral proteins separated under nonreducing conditions. (b) Immunoblot analysis of samples used for a with anti-HA2 antibodies reveals disulfide-bonded HA and HA fragments, separated by SDS/PAGE under nonreducing conditions. (c) Samples identical to those in a are separated by SDS/PAGE under reducing conditions and stained with Coomassie blue (lanes are as in a). (d) Immunoblot analysis of samples used for c with anti-HA2 antibodies reveals HA2 and HA2 fragments, separated by SDS/PAGE under reducing conditions (lanes are as in b). Identity of the HA2 fragment was confirmed for all four samples (pH 7.3, pH 5.0, 65°C, and 4.5 M urea) by five cycles of amino-terminal amino acid sequence analysis (see text). HA, an HA1 polypeptide disulfide bonded to an HA2 polypeptide. HA fragments, disulfide-bonded HA1 and HA2 proteolytic fragments. HA2, intact HA2 polypeptide. HA2 fragment, proteolytic fragment of HA2. M1, matrix protein. NP, nucleoprotein.
Figure 4
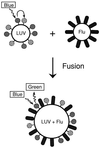
Schematic of the assay for membrane-fusion activity. Large, unilamellar vesicles (LUV) are prepared with fluorescent donor and acceptor lipids (solid and striped spheres, respectively). On excitation of the donor fluorophore (blue light, 465 nm), there is substantial resonance energy transfer to the acceptor fluorophore, so that little donor fluorescence is observed (green light, 535 nm). After membrane fusion, the lipids of the viral envelope (Flu) mix with the vesicle, separating the donor from the acceptor (LUV + Flu), leading to an increase in observed donor fluorescence.
Figure 8
A model for the mechanism of the HA conformational change. The metastable native state (N) undergoes an irreversible transition to the more stable fusogenic state (F). The fusogenic state induces membrane-fusion activity in the presence of target membranes and causes viral inactivation in the absence of target membranes.
Similar articles
- Inhibition of influenza virus hemagglutinin-mediated membrane fusion by a compound related to podocarpic acid.
Staschke KA, Hatch SD, Tang JC, Hornback WJ, Munroe JE, Colacino JM, Muesing MA. Staschke KA, et al. Virology. 1998 Sep 1;248(2):264-74. doi: 10.1006/viro.1998.9273. Virology. 1998. PMID: 9721235 - Probing the metastable state of influenza hemagglutinin.
Kingsley CN, Antanasijevic A, Palka-Hamblin H, Durst M, Ramirez B, Lavie A, Caffrey M. Kingsley CN, et al. J Biol Chem. 2017 Dec 29;292(52):21590-21597. doi: 10.1074/jbc.M117.815043. Epub 2017 Nov 10. J Biol Chem. 2017. PMID: 29127198 Free PMC article. - Hydrostatic pressure induces the fusion-active state of enveloped viruses.
Gaspar LP, Silva AC, Gomes AM, Freitas MS, Ano Bom AP, Schwarcz WD, Mestecky J, Novak MJ, Foguel D, Silva JL. Gaspar LP, et al. J Biol Chem. 2002 Mar 8;277(10):8433-9. doi: 10.1074/jbc.M106096200. Epub 2001 Nov 26. J Biol Chem. 2002. PMID: 11723114 - Early steps of the conformational change of influenza virus hemagglutinin to a fusion active state: stability and energetics of the hemagglutinin.
Huang Q, Sivaramakrishna RP, Ludwig K, Korte T, Böttcher C, Herrmann A. Huang Q, et al. Biochim Biophys Acta. 2003 Jul 11;1614(1):3-13. doi: 10.1016/s0005-2736(03)00158-5. Biochim Biophys Acta. 2003. PMID: 12873761 Review. - Influenza fusion peptides.
Skehel JJ, Cross K, Steinhauer D, Wiley DC. Skehel JJ, et al. Biochem Soc Trans. 2001 Aug;29(Pt 4):623-6. doi: 10.1042/bst0290623. Biochem Soc Trans. 2001. PMID: 11498040 Review.
Cited by
- Dynamic changes during acid-induced activation of influenza hemagglutinin.
Garcia NK, Guttman M, Ebner JL, Lee KK. Garcia NK, et al. Structure. 2015 Apr 7;23(4):665-76. doi: 10.1016/j.str.2015.02.006. Epub 2015 Mar 12. Structure. 2015. PMID: 25773144 Free PMC article. - Structures and polymorphic interactions of two heptad-repeat regions of the SARS virus S2 protein.
Deng Y, Liu J, Zheng Q, Yong W, Lu M. Deng Y, et al. Structure. 2006 May;14(5):889-99. doi: 10.1016/j.str.2006.03.007. Structure. 2006. PMID: 16698550 Free PMC article. - Delay of influenza hemagglutinin refolding into a fusion-competent conformation by receptor binding: a hypothesis.
Leikina E, Markovic I, Chernomordik LV, Kozlov MM. Leikina E, et al. Biophys J. 2000 Sep;79(3):1415-27. doi: 10.1016/S0006-3495(00)76393-4. Biophys J. 2000. PMID: 10969003 Free PMC article. - Bypassing the kinetic trap of serpin protein folding by loop extension.
Im H, Ahn HY, Yu MH. Im H, et al. Protein Sci. 2000 Aug;9(8):1497-502. doi: 10.1110/ps.9.8.1497. Protein Sci. 2000. PMID: 10975571 Free PMC article.
References
- Wiley D C, Skehel J J. Annu Rev Biochem. 1987;56:365–394. - PubMed
- Schoch C, Blumenthal R, Clague M J. FEBS Lett. 1992;311:221–225. - PubMed
- Stegmann T, Helenius A. In: Viral Fusion Mechanisms. Bentz J, editor. Boca Raton, FL: CRC; 1993. pp. 89–111.
- Hughson F M. Curr Opin Struct Biol. 1995;5:507–513. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources