Ancient mtDNA sequences in the human nuclear genome: a potential source of errors in identifying pathogenic mutations - PubMed (original) (raw)
Ancient mtDNA sequences in the human nuclear genome: a potential source of errors in identifying pathogenic mutations
D C Wallace et al. Proc Natl Acad Sci U S A. 1997.
Abstract
Nuclear-localized mtDNA pseudogenes might explain a recent report describing a heteroplasmic mtDNA molecule containing five linked missense mutations dispersed over the contiguous mtDNA CO1 and CO2 genes in Alzheimer's disease (AD) patients. To test this hypothesis, we have used the PCR primers utilized in the original report to amplify CO1 and CO2 sequences from two independent rho degrees (mtDNA-less) cell lines. CO1 and CO2 sequences amplified from both of the rho degrees cells, demonstrating that these sequences are also present in the human nuclear DNA. The nuclear pseudogene CO1 and CO2 sequences were then tested for each of the five "AD" missense mutations by restriction endonuclease site variant assays. All five mutations were found in the nuclear CO1 and CO2 PCR products from rho degrees cells, but none were found in the PCR products obtained from cells with normal mtDNA. Moreover, when the overlapping nuclear CO1 and CO2 PCR products were cloned and sequenced, all five missense mutations were found, as well as a linked synonymous mutation. Unlike the findings in the original report, an additional 32 base substitutions were found, including two in adjacent tRNAs and a two base pair deletion in the CO2 gene. Phylogenetic analysis of the nuclear CO1 and CO2 sequences revealed that they diverged from modern human mtDNAs early in hominid evolution about 770,000 years before present. These data would be consistent with the interpretation that the missense mutations proposed to cause AD may be the product of ancient mtDNA variants preserved as nuclear pseudogenes.
Figures
Figure 1
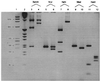
Amplification of PCR products from total cellular DNAs extracted from two different human cell lines and their mtDNA-deficient ρ° counterparts using various pairs of primers homologous to the normal human mtDNA sequence (26). The cell lines are 143B-TK− (lanes 3, 8, 13, and 18), 143B-87 (ρ°) (lanes 4, 9, 14, and 19), WAL2A (lanes 5, 10, 15, and 20), and WAL2A-EB2 (ρ°) (lanes 6, 11, 16, and 21). The primer pairs used are 14430FOR/15461REV (lanes 2–6) and 15238FOR/15865REV (lanes 7–11), both encompassing positions of cytochrome b; 5803COIFOR/7570 CO1REV encompassing CO1 (lanes 12–16) and 7483CO2FOR/8383CO2REV encompassing CO2 (lanes 17–21). Negative controls involving amplification without template are shown in lanes 2, 7, 12, and 17. A size standard is shown in lane 1.
Figure 2
Detection of the five missense mutations in CO1 and CO2 genes amplified from DNA of 143B-TK− and 143B-87 ρ° cells by restriction endonuclease digestion. PCR products obtained from 143B-TK− DNA are shown in lanes 3 and 5 for CO1 and lanes 7, 9, and 11 for CO2. PCR products obtained from 143B-87 ρ° DNA are shown in lanes 4 and 6 for CO1 and lanes 8, 10, and 12 for CO2. Detection of the np 6366 G-to-A mutation in CO1 by_Bsm_AI digestion is presented in lanes 3 and 4. Detection of the np 7146 A-to-G mutation in CO1 by _Nru_I digestion is shown in lanes 5 and 6. The double band in lane 6 is most likely the result of partial digestion. Detection of the np 7650 C-to-T mutation by _Hph_I digestion is shown in lanes 7 and 8. Detection of the np 7868 C-to-T mutation by _Mse_I digestion is shown in lanes 9 and 10. Detection of the np 8021 A-to-G mutations by_Hpa_II digestion is shown in lanes 11 and 12. Lanes 1 and 2 are size standards: lane 1 is a ∅X174 _Hae_III digest and lane 2 is a 1-kb DNA ladder.
Figure 3
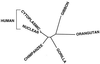
Unrooted NJ tree relating the human nuclear CO1 and CO2 sequences to the homologous sequences from the human, chimpanzee, gorilla, gibbon, and orangutan mtDNAs. This tree is based on genetic distances calculated using the maximum likelihood model (DNAML) (28). Boostrap analysis from 100 independent trees generated the diagrammed result in 100% of the comparisons between the human nuclear and cytoplasmic sequences and between the gibbon and orangutan sequences, in 99% of the comparisons between the two human sequences and the chimpanzee sequence, and in 82% of the comparisons between the chimpanzee and gorilla sequences and between the gorilla and the collective gibbon and orangutan sequences. Phylogenies with identical branching orders were obtained by using genetic distances calculated by the parsimony, Jukes-Cantor, and Kimura two-parameter methods.
Similar articles
- Evidence that two reports of mtDNA cytochrome c oxidase "mutations" in Alzheimer's disease are based on nDNA pseudogenes of recent evolutionary origin.
Davis JN 2nd, Parker WD Jr. Davis JN 2nd, et al. Biochem Biophys Res Commun. 1998 Mar 27;244(3):877-83. doi: 10.1006/bbrc.1998.8353. Biochem Biophys Res Commun. 1998. PMID: 9535760 - A novel mitochondrial DNA-like sequence in the human nuclear genome.
Herrnstadt C, Clevenger W, Ghosh SS, Anderson C, Fahy E, Miller S, Howell N, Davis RE. Herrnstadt C, et al. Genomics. 1999 Aug 15;60(1):67-77. doi: 10.1006/geno.1999.5907. Genomics. 1999. PMID: 10458912 - Mitochondrial pseudogenes in the nuclear genome of Aedes aegypti mosquitoes: implications for past and future population genetic studies.
Hlaing T, Tun-Lin W, Somboon P, Socheat D, Setha T, Min S, Chang MS, Walton C. Hlaing T, et al. BMC Genet. 2009 Mar 6;10:11. doi: 10.1186/1471-2156-10-11. BMC Genet. 2009. PMID: 19267896 Free PMC article. - Mitochondrial genomic contribution to mitochondrial dysfunction in Alzheimer's disease.
Onyango I, Khan S, Miller B, Swerdlow R, Trimmer P, Bennett P Jr. Onyango I, et al. J Alzheimers Dis. 2006 Jul;9(2):183-93. doi: 10.3233/jad-2006-9210. J Alzheimers Dis. 2006. PMID: 16873965 Review. - Genetic basis of Alzheimer's dementia: role of mtDNA mutations.
Grazina M, Pratas J, Silva F, Oliveira S, Santana I, Oliveira C. Grazina M, et al. Genes Brain Behav. 2006;5 Suppl 2:92-107. doi: 10.1111/j.1601-183X.2006.00225.x. Genes Brain Behav. 2006. PMID: 16681804 Review.
Cited by
- Mitochondrial DNA oxidative damage and repair in aging and Alzheimer's disease.
Santos RX, Correia SC, Zhu X, Smith MA, Moreira PI, Castellani RJ, Nunomura A, Perry G. Santos RX, et al. Antioxid Redox Signal. 2013 Jun 20;18(18):2444-57. doi: 10.1089/ars.2012.5039. Epub 2012 Dec 7. Antioxid Redox Signal. 2013. PMID: 23216311 Free PMC article. Review. - Dysregulation of circular RNAs in inflammation and cancers.
Liu J, Zhao F, Chen LL, Su S. Liu J, et al. Fundam Res. 2023 Jul 2;3(5):683-691. doi: 10.1016/j.fmre.2023.04.019. eCollection 2023 Sep. Fundam Res. 2023. PMID: 38933304 Free PMC article. Review. - Alzheimer's disease: diverse aspects of mitochondrial malfunctioning.
Santos RX, Correia SC, Wang X, Perry G, Smith MA, Moreira PI, Zhu X. Santos RX, et al. Int J Clin Exp Pathol. 2010 Jun 25;3(6):570-81. Int J Clin Exp Pathol. 2010. PMID: 20661404 Free PMC article. Review. - Identification of Somatic Mitochondrial DNA Mutations, Heteroplasmy, and Increased Levels of Catenanes in Tumor Specimens Obtained from Three Endometrial Cancer Patients.
Young MJ, Sachidanandam R, Hales DB, Brard L, Robinson K, Rahman MM, Khadka P, Groesch K, Young CKJ. Young MJ, et al. Life (Basel). 2022 Apr 9;12(4):562. doi: 10.3390/life12040562. Life (Basel). 2022. PMID: 35455053 Free PMC article. - PHFinder: assisted detection of point heteroplasmy in Sanger sequencing chromatograms.
Suárez Menéndez M, Rivera-León VE, Robbins J, Berube M, Palsbøll PJ. Suárez Menéndez M, et al. PeerJ. 2023 Sep 19;11:e16028. doi: 10.7717/peerj.16028. eCollection 2023. PeerJ. 2023. PMID: 37744223 Free PMC article.
References
- Wallace D C, Singh G, Lott M T, Hodge J A, Schurr T G, Lezza A M S, Elsas L J, II, Nikoskelainen E K. Science. 1988;242:1427–1430. - PubMed
- Wallace D C, Lott M T, Brown M D. In: Human Gene Mapping 1995. Cuticchia J, editor. Baltimore: Johns Hopkins Univ. Press; 1996. pp. 1280–1331.
- Wallace D C, Brown M D, Lott M T. In: Emery & Rimoin’s Principle & Practice of Medical Genetics. 3rd Ed. Rimoin D L, Connor J M, Pyeritz R E, editors. New York: Churchill Livingstone; 1996. pp. 277–332.
- Wallace D C. In: The Molecular & Genetic Basis of Neurological Disease. Rosenberg R N, Prusiner S B, DiMauro S, Barchi R L, editors. Boston: Butterworth-Heinemann; 1997. pp. 237–269.
Publication types
MeSH terms
Substances
Grants and funding
- R01 NS021328/NS/NINDS NIH HHS/United States
- HL45572/HL/NHLBI NIH HHS/United States
- NS21328/NS/NINDS NIH HHS/United States
- R01 AG013154/AG/NIA NIH HHS/United States
- AG13154/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical