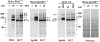Evidence for covalent modification of the nuclear dot-associated proteins PML and Sp100 by PIC1/SUMO-1 - PubMed (original) (raw)
Evidence for covalent modification of the nuclear dot-associated proteins PML and Sp100 by PIC1/SUMO-1
T Sternsdorf et al. J Cell Biol. 1997.
Abstract
PML and Sp100 proteins are associated with nuclear domains, known as nuclear dots (NDs). They were discovered in the context of leukemic transformation and as an autoantigen in primary biliary cirrhosis, respectively. Both proteins are expressed in the form of many COOH-terminally spliced variants, and their expression is enhanced by interferons (IFN). The recent finding that PIC1/SUMO-1, a small ubiquitin-like protein, is covalently linked to the RanGAP1 protein of the nuclear pore complex and also binds PML in yeast cells led us to determine whether PML is covalently modified by PIC1/SUMO-1 and whether the same is true for Sp100. We found an immune reaction of PML and Sp100 proteins with a PIC1/SUMO-1-specific monoclonal antibody by immunoblotting when using cell extracts prepared from stably transfected cells inducibly expressing one isoform of each protein as well as from nontransfected cells. In contrast, both proteins did not react when synthesized in vitro. Immunofluorescence staining showed that PIC1/SUMO-1 colocalized with Sp100 and PML in NDs except in mitotic cells, in which PML and Sp100 are dissociated. Cell fractionation and immunoblotting demonstrated that PIC1/SUMO-1 immunoreactive Sp100 in IFN-treated and untreated cells was exclusively nuclear, whereas nonmodified Sp100 was also found in the cytoplasm. Taken together, these data strongly suggest covalent modification of specific nuclear isoforms of Sp100 and PML by PIC1/SUMO-1. This modification may play a regulatory role in ND structure, composition, and function.
Figures
Figure 1
Schematic representation of sequence motivs in the Sp100 and PML splice variants used. Sp100 represents the originally described major splice variant (Szostecki et al., 1990). In its NH2-terminal region, Sp100 harbors a region with homology to MHC class I molecules, followed by a domain that is conserved in its murine homologue (Grötzinger et al., 1996_a_; Weichenhan et al., 1997) and in a lymphocyte-specific ND protein (Bloch et al., 1996; Dent et al., 1996), termed HSR domain (Sternsdorf et al., 1997_a_). Also depicted is a region of sequence homology to the HIV I Nef-protein. The COOH terminus of Sp100 is acidic and contains a consensus phosphorylation site for casein kinase 2. This region (32 amino acids) is replaced by a basic stretch of 24 amino acids in SpAlt-C (Guldner, H., C. Szostecki, and H. Will, manuscript submitted for publication). The PML splice variant used in this study contains the proline-rich NH2 terminus, the RING-finger-B-Box motiv, followed by the predicted coiled coil region (de Thé et al., 1991). At its COOH terminus, the PML protein contains the nuclear localization signal (NLS), a serine-rich region, and five repeats of the amino acid motiv LASP(L).
Figure 2
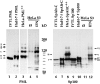
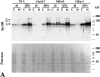
Immunoblot analysis of in vitro translated PML, SpAlt-C, and the Sp100 protein as well as of proteins of total cell extracts from various cells (HuH-7 cells transfected with PML, SpAlt-C and Sp100 expression vectors, induced HeLa-PML++ and HeLa-SpAltC++ cells, and HeLa S3 cells with and without prior treatment with IFN) separated by 7.5% SDS-PAGE. PML protein synthesized in vitro (lane 1, IVTL PML) has a higher electrophoretic mobility (band A) than the same PML protein expressed in transiently transfected HuH-7 (lane 2, band B) or in HeLa-PML++ cells (lane 3, band B), the latter showing additional bands of lower electrophoretic mobility (lane 3, bands C–F). PML proteins expressed from the endogenous gene are visualized as multiple bands in IFN-treated HeLa S3 cells (lane 5). In untreated cells, only the most prominent one is observed (lane 4). SpAlt-C protein comigrates when synthesized in vitro or in HuH-7 cells (lanes 6 and 7, band I). When expressed in HeLa-SpAltC++ cells (lane 8, band I), an additional band corresponding to a 110-kD protein is visualized (lane 8, band II). The Sp100 protein translated in vitro differs from that expressed transiently in HuH-7 cells by ∼3 kD (lane 10, bands III and IV). Sp100 proteins expressed from the endogenous gene in IFN-treated HeLa S3 cells are visualized as multiple bands; the major ones are indicated (lane 12, bands IV–VI). In untreated HeLa S3 cells, little Sp100 protein is detected (lane 11).
Figure 3
Immunoblot with proteins of total extracts from noninduced (lanes + TET) and induced (lanes Ø TET) HeLa-PML++ and HeLa-SpAltC++ cells, and of untreated (lanes Ø) and IFN-treated (lanes IFN) HeLa S3 cells, separated by 12.5% SDS-PAGE. For detection, blots were incubated with polyclonal rat-anti-PML (PML), rat-anti-Sp100 (Sp100) Abs, and PIC1/SUMO-1–specific mAb 21C7 (21C7). In induced HeLa-PML++ cells, several strong bands were stained with anti-PML Abs (bands B–F) but only some of them (bands C–F) with anti-PIC1/SUMO-1 mAb. This indicates covalent linkage of PIC1/ SUMO-1 or a related polypeptide to proteins in the latter bands. In induced HeLa-SpAltC++, two major bands emerged when stained with anti-Sp100 Abs (bands I and II), whereas only the protein in band II, but not in band I, reacted with mAb 21C7. This indicates that band II represents the PIC1/SUMO-1–like modified form of the protein present in band I. In total extracts from untreated and IFN-treated HeLa S3 cells, a minor increase in staining by mAb 21C7 after IFN treatment in the range of 100–200 kD was observed. In all lanes, a 90-kD protein was recognized (band RG), which represents PIC1/SUMO-1–modified RanGAP1 protein (compare Fig. 4). Staining of the same blots with Ponceau S shows similar amounts of protein in all lanes (right; Ponceau S, shown for HeLa-SpAltC++).
Figure 4
Detection of endogenous Sp100 carrying the PIC1/ SUMO1–like modification after immunoprecipitation from total nuclear extracts of HuH-7 cells with rabbit anti-Sp100 Abs (SpDF). Immunoblot of total nuclear extracts (lanes NE total), supernatants (lanes SN), and precipitates (lanes IP) from untreated (lanes Ø) and IFN-treated (lanes IFN) cells, separated on a 7.5% SDS gel. Blots were incubated with rat anti-Sp100 Abs, mAb 19C7 (specific for RanGAP1), 21C7 (specific for PIC1/ SUMO-1), or Ubi-1 (specific for ubiquitin). After immunoprecipitation, Sp100 proteins are detected in the precipitate of IFN-treated cells but hardly in the precipitate of untreated cells or in the supernatant. When incubated with mAb 21C7, a band corresponding to Sp100 band V became visible only in the precipitate of IFN-treated cells but not of untreated cells, indicating that band V corresponds to a PIC1/SUMO-1–modified form of Sp100. A large number of bands became visible in the supernatant, with the most prominent band at a relative mobility of 90 kD. Control staining of a parallel blot with mAb 19C7 confirms that this band represents the PIC1/SUMO-1–modified form of the RanGAP protein (shown for supernatant of IFN-treated cells; position of RanGAP marked by asterisk). A blot with the same extracts stained with a ubiquitin-specific mAb showed no detectable reactive protein in the precipitate, even after long exposure of the blot. In contrast, many bands corresponding to ubiquitinylated cellular proteins were observed in the supernatant.
Figure 5
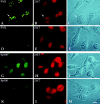
Immunofluorescence staining of induced and noninduced HeLa-SpAltC++ and HeLa-PML++ cells fixed with methanol/acetone. In induced HeLa-PML++ cells, rat anti-PML Ab shows bright and enlarged ND staining in nonmitotic cells and large aggregates in a mitotic cell (A, arrow), whereas only a few small dots were stained in the noninduced cells (D). Double staining of the same slides with mAb 21C7 shows overlapping ND staining in induced nonmitotic cells (D) and a diffuse homogeneous pattern in noninduced cells (E). In mitosis, induced HeLa-PML++ cells resulted in no staining of PML aggregates with mAb 21C7 (B, arrow). For comparison, phase contrast images are given in C and F. In induced HeLa-SpAltC++ cells, rabbit anti-Sp100 Abs resulted in a microspeckled pattern with some large dots (G), whereas only a few small dots were stained in the noninduced cells (K). Additional incubation of the same slides with mAb 21C7 led to homogenous staining both in induced (H) and noninduced cells (L). For comparison, phase contrast images are given in I and M. Bar, 10 μm.
Figure 6
Immunofluorescence staining of methanol/ acetone-fixed, IFN-treated HeLa S3 cells. Cells were either double-labeled with polyclonal rat anti-PML (green) and mAb 21C7 (red) (A–C), with polyclonal rabbit anti-Sp100 antibodies (green) and mAb 21C7 (red) (D to F), or with anti-PML Abs and anti-Sp100 Abs (G–I). In a mitotic cell, PML is aggregated at the periphery of the cell (A and G, arrow), whereas Sp100 and PIC1/ SUMO-1 are diffusely distributed (B, D, E, and H), indicating that PIC1/SUMO-1 and Sp100 dissociate from PML in mitosis. Bar, 10 μm.
Figure 7
(A) Differentiation of nuclear and cytoplasmic forms of Sp100 proteins from various untreated (Ø) and IFN-treated (IFN) cell lines on immunoblots (12.5% SDS-PAGE) incubated with anti-Sp100 antibodies. With the exception of HuH-7, an Sp100 protein of ∼88 kD (band IV) is detected not only in the nuclear (lanes N) but also in the cytoplasmic fractions (lanes C) of all cell lines tested. In untreated HuH-7 cells, this protein is detected exclusively in the cytoplasmic fraction. With the exception of MG63 cells, IFN treatment strongly increases Sp100 levels in the nuclear fractions and only moderately in the cytoplasmic fractions. Isoforms of Sp100 with slower electrophoretic mobility (bands V–VII, arrow) are detectable only in the nuclear fractions. A similar amount of proteins of the respective fractions was loaded as documented by Ponceau S staining (Ponceau). (B) Immunoblot with nuclear and cytoplasmic extracts of HuH-7 and HEp-2 cells and polyclonal rabbit anti-HNF-1 (HNF-1) and monoclonal anti-Sp1 (Sp1) antibodies, and a human autoimmune serum, recognizing among other proteins the mitochondrial 74-kD dihydrolipoamide acetyltransferase subunit (p74). The transcription factors HNF-1 and Sp1 are detected predominantly in the nuclear fraction (lanes N) of HuH-7 or HuH-7 and HEp-2, cells, respectively (arrows). In contrast, the 74-kD mitochondrial protein was detected exclusively in the cytoplasmic fraction of HEp-2 cells (p74, lanes C, arrow). Treatment of the cells with interferon does not alter the subcellular localization of these proteins (lanes IFN). Lanes LM, Ponceau S staining of the molecular mass marker of the corresponding blots. (C) Differentiation of nuclear and cytoplasmic forms of Sp100 proteins in untreated (Ø) and IFN-treated (IFN) HeLa S3 cells, in induced (ØTet) and noninduced (+Tet) HeLa-PML++ as well as in HeLa-SpAlt-C++ cells by immunoblotting (12.5% SDS-PAGE) with anti-Sp100 or anti-PML antibodies. Similar to A, Sp100 as well as PML proteins are both in the nuclear (lanes N) and cytoplasmic (lanes C) fractions. IFN treatment as well as induction of HeLa-PML++ or HeLa-SpAltC++ cells increase their amounts predominantly in the nuclear fraction. Modified Sp100 proteins (bands II, V, and VI) are exclusively nuclear. Modified PML protein also is detected exclusively in nuclei of HeLa-PML++ cells. This is not evident in nontransfected HeLa S3 cells. Little Sp100 and PML proteins are detected in a nuclear wash fraction (lanes W).
Figure 7
(A) Differentiation of nuclear and cytoplasmic forms of Sp100 proteins from various untreated (Ø) and IFN-treated (IFN) cell lines on immunoblots (12.5% SDS-PAGE) incubated with anti-Sp100 antibodies. With the exception of HuH-7, an Sp100 protein of ∼88 kD (band IV) is detected not only in the nuclear (lanes N) but also in the cytoplasmic fractions (lanes C) of all cell lines tested. In untreated HuH-7 cells, this protein is detected exclusively in the cytoplasmic fraction. With the exception of MG63 cells, IFN treatment strongly increases Sp100 levels in the nuclear fractions and only moderately in the cytoplasmic fractions. Isoforms of Sp100 with slower electrophoretic mobility (bands V–VII, arrow) are detectable only in the nuclear fractions. A similar amount of proteins of the respective fractions was loaded as documented by Ponceau S staining (Ponceau). (B) Immunoblot with nuclear and cytoplasmic extracts of HuH-7 and HEp-2 cells and polyclonal rabbit anti-HNF-1 (HNF-1) and monoclonal anti-Sp1 (Sp1) antibodies, and a human autoimmune serum, recognizing among other proteins the mitochondrial 74-kD dihydrolipoamide acetyltransferase subunit (p74). The transcription factors HNF-1 and Sp1 are detected predominantly in the nuclear fraction (lanes N) of HuH-7 or HuH-7 and HEp-2, cells, respectively (arrows). In contrast, the 74-kD mitochondrial protein was detected exclusively in the cytoplasmic fraction of HEp-2 cells (p74, lanes C, arrow). Treatment of the cells with interferon does not alter the subcellular localization of these proteins (lanes IFN). Lanes LM, Ponceau S staining of the molecular mass marker of the corresponding blots. (C) Differentiation of nuclear and cytoplasmic forms of Sp100 proteins in untreated (Ø) and IFN-treated (IFN) HeLa S3 cells, in induced (ØTet) and noninduced (+Tet) HeLa-PML++ as well as in HeLa-SpAlt-C++ cells by immunoblotting (12.5% SDS-PAGE) with anti-Sp100 or anti-PML antibodies. Similar to A, Sp100 as well as PML proteins are both in the nuclear (lanes N) and cytoplasmic (lanes C) fractions. IFN treatment as well as induction of HeLa-PML++ or HeLa-SpAltC++ cells increase their amounts predominantly in the nuclear fraction. Modified Sp100 proteins (bands II, V, and VI) are exclusively nuclear. Modified PML protein also is detected exclusively in nuclei of HeLa-PML++ cells. This is not evident in nontransfected HeLa S3 cells. Little Sp100 and PML proteins are detected in a nuclear wash fraction (lanes W).
Figure 7
(A) Differentiation of nuclear and cytoplasmic forms of Sp100 proteins from various untreated (Ø) and IFN-treated (IFN) cell lines on immunoblots (12.5% SDS-PAGE) incubated with anti-Sp100 antibodies. With the exception of HuH-7, an Sp100 protein of ∼88 kD (band IV) is detected not only in the nuclear (lanes N) but also in the cytoplasmic fractions (lanes C) of all cell lines tested. In untreated HuH-7 cells, this protein is detected exclusively in the cytoplasmic fraction. With the exception of MG63 cells, IFN treatment strongly increases Sp100 levels in the nuclear fractions and only moderately in the cytoplasmic fractions. Isoforms of Sp100 with slower electrophoretic mobility (bands V–VII, arrow) are detectable only in the nuclear fractions. A similar amount of proteins of the respective fractions was loaded as documented by Ponceau S staining (Ponceau). (B) Immunoblot with nuclear and cytoplasmic extracts of HuH-7 and HEp-2 cells and polyclonal rabbit anti-HNF-1 (HNF-1) and monoclonal anti-Sp1 (Sp1) antibodies, and a human autoimmune serum, recognizing among other proteins the mitochondrial 74-kD dihydrolipoamide acetyltransferase subunit (p74). The transcription factors HNF-1 and Sp1 are detected predominantly in the nuclear fraction (lanes N) of HuH-7 or HuH-7 and HEp-2, cells, respectively (arrows). In contrast, the 74-kD mitochondrial protein was detected exclusively in the cytoplasmic fraction of HEp-2 cells (p74, lanes C, arrow). Treatment of the cells with interferon does not alter the subcellular localization of these proteins (lanes IFN). Lanes LM, Ponceau S staining of the molecular mass marker of the corresponding blots. (C) Differentiation of nuclear and cytoplasmic forms of Sp100 proteins in untreated (Ø) and IFN-treated (IFN) HeLa S3 cells, in induced (ØTet) and noninduced (+Tet) HeLa-PML++ as well as in HeLa-SpAlt-C++ cells by immunoblotting (12.5% SDS-PAGE) with anti-Sp100 or anti-PML antibodies. Similar to A, Sp100 as well as PML proteins are both in the nuclear (lanes N) and cytoplasmic (lanes C) fractions. IFN treatment as well as induction of HeLa-PML++ or HeLa-SpAltC++ cells increase their amounts predominantly in the nuclear fraction. Modified Sp100 proteins (bands II, V, and VI) are exclusively nuclear. Modified PML protein also is detected exclusively in nuclei of HeLa-PML++ cells. This is not evident in nontransfected HeLa S3 cells. Little Sp100 and PML proteins are detected in a nuclear wash fraction (lanes W).
Figure 8
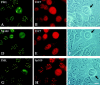
Detection of cytoplasmic Sp100 protein in a coculture of human HuH-7 and rat R1H cells by immunofluorescence analysis. Untreated HuH-7 cells when stained with polyclonal rabbit anti-Sp100 Abs (Sp26) lack ND staining but show cytoplasmic fluorescence (Sp100/Ø). After treatment with IFN, a clear ND staining became visible in addition to the cytoplasmic staining (Sp100/IFN). Both images were taken at the same exposure time. At this exposure, rat R1H cells are not visibly stained by the Abs and serve as an internal negative control. Localization of the rat cells is visible in the corresponding phase contrast pictures given below. Bar, 10 μm.
Similar articles
- Cellular localization, expression, and structure of the nuclear dot protein 52.
Sternsdorf T, Jensen K, Züchner D, Will H. Sternsdorf T, et al. J Cell Biol. 1997 Jul 28;138(2):435-48. doi: 10.1083/jcb.138.2.435. J Cell Biol. 1997. PMID: 9230084 Free PMC article. - Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins.
Chelbi-Alix MK, de Thé H. Chelbi-Alix MK, et al. Oncogene. 1999 Jan 28;18(4):935-41. doi: 10.1038/sj.onc.1202366. Oncogene. 1999. PMID: 10023669 - Small ubiquitin-related modifiers: A novel and independent class of autoantigens in primary biliary cirrhosis.
Janka C, Selmi C, Gershwin ME, Will H, Sternsdorf T. Janka C, et al. Hepatology. 2005 Mar;41(3):609-16. doi: 10.1002/hep.20619. Hepatology. 2005. PMID: 15726652 - Autoantibodies against "nuclear dots" in primary biliary cirrhosis.
Szostecki C, Guldner HH, Will H. Szostecki C, et al. Semin Liver Dis. 1997 Feb;17(1):71-8. doi: 10.1055/s-2007-1007184. Semin Liver Dis. 1997. PMID: 9089912 Review. - Nuclear dots: actors on many stages.
Sternsdorf T, Grötzinger T, Jensen K, Will H. Sternsdorf T, et al. Immunobiology. 1997 Dec;198(1-3):307-31. doi: 10.1016/S0171-2985(97)80051-4. Immunobiology. 1997. PMID: 9442402 Review.
Cited by
- The potential link between PML NBs and ICP0 in regulating lytic and latent infection of HSV-1.
Wang S, Long J, Zheng CF. Wang S, et al. Protein Cell. 2012 May;3(5):372-82. doi: 10.1007/s13238-012-2021-x. Epub 2012 Apr 28. Protein Cell. 2012. PMID: 22544561 Free PMC article. Review. - Detecting endogenous SUMO targets in mammalian cells and tissues.
Becker J, Barysch SV, Karaca S, Dittner C, Hsiao HH, Berriel Diaz M, Herzig S, Urlaub H, Melchior F. Becker J, et al. Nat Struct Mol Biol. 2013 Apr;20(4):525-31. doi: 10.1038/nsmb.2526. Epub 2013 Mar 17. Nat Struct Mol Biol. 2013. PMID: 23503365 - Interaction between neuronal intranuclear inclusions and promyelocytic leukemia protein nuclear and coiled bodies in CAG repeat diseases.
Yamada M, Sato T, Shimohata T, Hayashi S, Igarashi S, Tsuji S, Takahashi H. Yamada M, et al. Am J Pathol. 2001 Nov;159(5):1785-95. doi: 10.1016/S0002-9440(10)63025-8. Am J Pathol. 2001. PMID: 11696439 Free PMC article. - Covalent modification of the transcriptional repressor tramtrack by the ubiquitin-related protein Smt3 in Drosophila flies.
Lehembre F, Badenhorst P, Müller S, Travers A, Schweisguth F, Dejean A. Lehembre F, et al. Mol Cell Biol. 2000 Feb;20(3):1072-82. doi: 10.1128/MCB.20.3.1072-1082.2000. Mol Cell Biol. 2000. PMID: 10629064 Free PMC article. - Evidence for a dual antiviral role of the major nuclear domain 10 component Sp100 during the immediate-early and late phases of the human cytomegalovirus replication cycle.
Tavalai N, Adler M, Scherer M, Riedl Y, Stamminger T. Tavalai N, et al. J Virol. 2011 Sep;85(18):9447-58. doi: 10.1128/JVI.00870-11. Epub 2011 Jul 6. J Virol. 2011. PMID: 21734036 Free PMC article.
References
- Ahn M-J, Nason-Burchenal K, Moasser MM, Dmitrovsky E. Growth suppression of acute promyelocytic leukemia cells having increased expression of the non-rearranged alleles: RARα or PML. Oncogene. 1995;10:2307–2314. - PubMed
- Aso T, Lane WS, Conaway JW, Conaway RC. Elongin (SIII): a multisubunit regulator of elongation by RNA polymerase II. Science. 1995;269:1439–1443. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases