A distinct and parallel pathway for the nuclear import of an mRNA-binding protein - PubMed (original) (raw)
A distinct and parallel pathway for the nuclear import of an mRNA-binding protein
L F Pemberton et al. J Cell Biol. 1997.
Abstract
Three independent pathways of nuclear import have so far been identified in yeast, each mediated by cognate nuclear transport factors, or karyopherins. Here we have characterized a new pathway to the nucleus, mediated by Mtr10p, a protein first identified in a screen for strains defective in polyadenylated RNA export. Mtr10p is shown to be responsible for the nuclear import of the shuttling mRNA-binding protein Npl3p. A complex of Mtr10p and Npl3p was detected in cytosol, and deletion of Mtr10p was shown to lead to the mislocalization of nuclear Npl3p to the cytoplasm, correlating with a block in import. Mtr10p bound peptide repeat-containing nucleoporins and Ran, suggesting that this import pathway involves a docking step at the nuclear pore complex and is Ran dependent. This pathway of Npl3p import is distinct and does not appear to overlap with another known import pathway for an mRNA-binding protein. Thus, at least two parallel pathways function in the import of mRNA-binding proteins, suggesting the need for the coordination of these pathways.
Figures
Figure 1
Mtr10p shares similarity with Kap95p. Comparison of amino acid sequences of Kap95p and Mtr10p. The proteins are aligned using CLUSTAL v.1.6 (DNAStar, Madison, WI). Identical amino acids are boxed; similar amino acids are shaded.
Figure 2
Deletion of MTR10 causes severe growth defects and temperature sensitivity. Wild-type, the _MTR10_-deleted strain (mtr10::HIS3), a strain with a protein A–tagged copy of MTR10 (Mtr10-PrA), and the _MTR10_-deleted strain complemented with the MTR10 gene on a plasmid (mtr10:HIS3, pRS316-MTR10) were streaked on rich medium at 23 and 37°C.
Figure 3
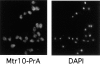
Mtr10p is primarily localized to the cytoplasm. The localization of Mtr10-PrA was examined by immunofluorescent detection of the protein A tag; the coincident staining of DNA with DAPI is shown.
Figure 4
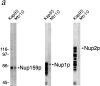
Mtr10p binds to a subset of repeat-containing nucleoporins. (a) Blot overlay assays were used to show binding of Mtr10-PrA to E. coli lysates containing repeat domain fragments of Nup159p (amino acids 441–876) and Nup1p (amino acids 432– 816) but not to bacterially expressed purified Nup2p. Blots were incubated with whole yeast cytosol from protein A–tagged strains; bound proteins were detected via the protein A moiety. Multiple bands detected with Kap95-PrA are due to its ability to bind strongly to proteolytic degradation fragments of the nucleoporin. Molecular masses in kD are shown. (b) The repeat domain (R; amino acids 441–876) and a nonrepeat-containing domain (NR; amino acids 176–440) of Nup159p were expressed in bacteria; the bacterial lysates were blotted as before. Overlay assays were carried out using a bacterial lysate containing GST-MTR10NT or unfused GST. Bound proteins were detected with a GST antiserum. Arrows indicate the position of Nup159N or Nup159NR on the blot. Molecular masses in kD are shown. (c) Kap95-PrA but not Mtr10-PrA interacts with bacterially expressed, purified Kap60p by overlay blot assay.
Figure 4
Mtr10p binds to a subset of repeat-containing nucleoporins. (a) Blot overlay assays were used to show binding of Mtr10-PrA to E. coli lysates containing repeat domain fragments of Nup159p (amino acids 441–876) and Nup1p (amino acids 432– 816) but not to bacterially expressed purified Nup2p. Blots were incubated with whole yeast cytosol from protein A–tagged strains; bound proteins were detected via the protein A moiety. Multiple bands detected with Kap95-PrA are due to its ability to bind strongly to proteolytic degradation fragments of the nucleoporin. Molecular masses in kD are shown. (b) The repeat domain (R; amino acids 441–876) and a nonrepeat-containing domain (NR; amino acids 176–440) of Nup159p were expressed in bacteria; the bacterial lysates were blotted as before. Overlay assays were carried out using a bacterial lysate containing GST-MTR10NT or unfused GST. Bound proteins were detected with a GST antiserum. Arrows indicate the position of Nup159N or Nup159NR on the blot. Molecular masses in kD are shown. (c) Kap95-PrA but not Mtr10-PrA interacts with bacterially expressed, purified Kap60p by overlay blot assay.
Figure 5
Mtr10p binds to Gsp1p. Bacterial lysates either induced (I) or uninduced (U) for GST-Mtr10NT, and bacterially expressed Kap95p were separated by SDS-PAGE and blotted. Blots were probed with Gsp1p-α[32P]GTP in an overlay assay, washed, and subjected to autoradiography. Bracket represents position of GST-Mtr10NT; additional bands are most likely degradation products of this protein. Molecular masses in kD are shown.
Figure 6
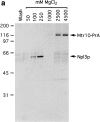
Immuno-isolation of a complex containing Mtr10-PrA and the mRNA-binding protein Npl3p. (a) Cytosol prepared from Mtr10-PrA cells was incubated with IgG sepharose; the Mtr10-PrA and complexed material were eluted in a MgCl2 step gradient. Fractions were analyzed by SDS-PAGE and Coomassie blue staining. The final wash fraction was also loaded (wash); numbers above lanes indicate molarity of MgCl2. The indicated band containing Npl3p was excised and analyzed by mass spectrometry. (b) The 250 mM eluate from a was immunoblotted with an antibody specific for Npl3p. Numbers to the left of both figures indicate molecular mass in kD.
Figure 6
Immuno-isolation of a complex containing Mtr10-PrA and the mRNA-binding protein Npl3p. (a) Cytosol prepared from Mtr10-PrA cells was incubated with IgG sepharose; the Mtr10-PrA and complexed material were eluted in a MgCl2 step gradient. Fractions were analyzed by SDS-PAGE and Coomassie blue staining. The final wash fraction was also loaded (wash); numbers above lanes indicate molarity of MgCl2. The indicated band containing Npl3p was excised and analyzed by mass spectrometry. (b) The 250 mM eluate from a was immunoblotted with an antibody specific for Npl3p. Numbers to the left of both figures indicate molecular mass in kD.
Figure 7
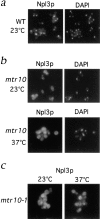
Npl3p is mislocalized to the cytoplasm in the absence of Mtr10p. (a) Immunofluorescent localization of Npl3p in wild-type (WT) cells colocalizes with the nucleus as visualized by DAPI-stained DNA. (b) Localization of Npl3p in the mtr10 deletion strain grown at 23 and 37°C shows that Npl3p is mislocalized to the cytoplasm when compared with the coincident DAPI staining. (c) Localization of Npl3p in the mtr10-1 mutant strain grown at 23 and 37°C shows that Npl3p is mislocalized to the cytoplasm when compared with the coincident DAPI staining.
Figure 8
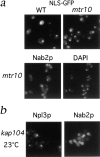
The Npl3p import pathway does not overlap with other import pathways. (a) Localization of an SV-40 NLS-GFP reporter was examined in living WT and mtr10 deletion cells and appeared nuclear in both cases. (b) Immunofluorescent localization of Nab2p in the mtr10 deletion strain shows correct nuclear localization. (c) Localization of Npl3p and Nab2p in a kap104:: HIS3 deletion strain at 23°C shows only mislocalization of Nab2p.
Similar articles
- Mtr10p functions as a nuclear import receptor for the mRNA-binding protein Npl3p.
Senger B, Simos G, Bischoff FR, Podtelejnikov A, Mann M, Hurt E. Senger B, et al. EMBO J. 1998 Apr 15;17(8):2196-207. doi: 10.1093/emboj/17.8.2196. EMBO J. 1998. PMID: 9545233 Free PMC article. - Uncoupling of the hnRNP Npl3p from mRNAs during the stress-induced block in mRNA export.
Krebber H, Taura T, Lee MS, Silver PA. Krebber H, et al. Genes Dev. 1999 Aug 1;13(15):1994-2004. doi: 10.1101/gad.13.15.1994. Genes Dev. 1999. PMID: 10444597 Free PMC article. - Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins.
Aitchison JD, Blobel G, Rout MP. Aitchison JD, et al. Science. 1996 Oct 25;274(5287):624-7. doi: 10.1126/science.274.5287.624. Science. 1996. PMID: 8849456 - Nucleocytoplasmic transport.
Görlich D, Mattaj IW. Görlich D, et al. Science. 1996 Mar 15;271(5255):1513-8. doi: 10.1126/science.271.5255.1513. Science. 1996. PMID: 8599106 Review. - Distinct nuclear import and export pathways mediated by members of the karyopherin beta family.
Moroianu J. Moroianu J. J Cell Biochem. 1998 Aug 1;70(2):231-9. J Cell Biochem. 1998. PMID: 9671229 Review.
Cited by
- Nuclear export of the small ribosomal subunit requires the ran-GTPase cycle and certain nucleoporins.
Moy TI, Silver PA. Moy TI, et al. Genes Dev. 1999 Aug 15;13(16):2118-33. doi: 10.1101/gad.13.16.2118. Genes Dev. 1999. PMID: 10465789 Free PMC article. - Autoregulation of Npl3, a yeast SR protein, requires a novel downstream region and serine phosphorylation.
Lund MK, Kress TL, Guthrie C. Lund MK, et al. Mol Cell Biol. 2008 Jun;28(11):3873-81. doi: 10.1128/MCB.02153-07. Epub 2008 Apr 7. Mol Cell Biol. 2008. PMID: 18391019 Free PMC article. - Crm1p mediates regulated nuclear export of a yeast AP-1-like transcription factor.
Yan C, Lee LH, Davis LI. Yan C, et al. EMBO J. 1998 Dec 15;17(24):7416-29. doi: 10.1093/emboj/17.24.7416. EMBO J. 1998. PMID: 9857197 Free PMC article. - A novel in vivo assay reveals inhibition of ribosomal nuclear export in ran-cycle and nucleoporin mutants.
Hurt E, Hannus S, Schmelzl B, Lau D, Tollervey D, Simos G. Hurt E, et al. J Cell Biol. 1999 Feb 8;144(3):389-401. doi: 10.1083/jcb.144.3.389. J Cell Biol. 1999. PMID: 9971735 Free PMC article. - A link between the synthesis of nucleoporins and the biogenesis of the nuclear envelope.
Marelli M, Lusk CP, Chan H, Aitchison JD, Wozniak RW. Marelli M, et al. J Cell Biol. 2001 May 14;153(4):709-24. doi: 10.1083/jcb.153.4.709. J Cell Biol. 2001. PMID: 11352933 Free PMC article.
References
- Aitchison JD, Blobel G, Rout MP. Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science. 1996;274:624–627. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous