A nuclear import pathway for a protein involved in tRNA maturation - PubMed (original) (raw)
A nuclear import pathway for a protein involved in tRNA maturation
J S Rosenblum et al. J Cell Biol. 1997.
Abstract
A limited number of transport factors, or karyopherins, ferry particular substrates between the cytoplasm and nucleoplasm. We identified the Saccharomyces cerevisiae gene YDR395w/SXM1 as a potential karyopherin on the basis of limited sequence similarity to known karyopherins. From yeast cytosol, we isolated Sxm1p in complex with several potential import substrates. These substrates included Lhp1p, the yeast homologue of the human autoantigen La that has recently been shown to facilitate maturation of pre-tRNA, and three distinct ribosomal proteins, Rpl16p, Rpl25p, and Rpl34p. Further, we demonstrate that Lhp1p is specifically imported by Sxm1p. In the absence of Sxm1p, Lhp1p was mislocalized to the cytoplasm. Sxm1p and Lhp1p represent the karyopherin and a cognate substrate of a unique nuclear import pathway, one that operates upstream of a major pathway of pre-tRNA maturation, which itself is upstream of tRNA export in wild-type cells. In addition, through its association with ribosomal proteins, Sxm1p may have a role in coordinating ribosome biogenesis with tRNA processing.
Figures
Figure 5
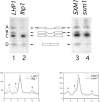
3′ processing of pre-tRNA Ser CGA Ser CGA in _LHP1_- and _SXM1_-deletion strains. A schematic of the intron-containing pre-tRNAs, species A, B, and C and of mature tRNA, species D, is included for reference. Exons are represented by boxes, 5′ and 3′ extensions are represented by black bars, and the central intron is shown. A Northern blot of total RNA was probed for intermediates A, B, and C using an oligonucleotide from the intron, and for mature tRNA using a probe that spans the properly spliced exons. Lane 1, wild-type cells (LHP1); lane 2, lhp1 cells; lane 3, Lhp1-PrA cells, wild-type for SXM1 (SXM1); and lane 4, Lhp1-PrA/sxm1 cells (sxm1). The blot was quantified using a PhosphorImager with ImageQuant software (Molecular Dynamics, Sunnyvale, CA). The graphs indicate radioactivity relative to distance, and peaks corresponding to intermediates are labeled. To account for minor loading discrepancies, the data in the graphs have been normalized to an internal control, precursor C, which was shown previously (Yoo and Wolin, 1997) to be independent of LHP1 genotype.
Figure 1
Kap95p and Sxm1p are homologous. The sequences were aligned by the Clustal method in MegAlign (DNA Star, Inc., Madison, WI). Similar amino acids are shaded, and identical amino acids are boxed and shaded.
Figure 2
Sxm1–PrA is localized throughout the cell. The PrA moiety was visualized by indirect immunofluorescence. For reference, the right panel represents the nuclei of the same field of cells by staining for DNA with DAPI.
Figure 3
Immunoisolation of cytosolic Sxm1–PrA interacting proteins. Whole cytosol from an Sxm1–PrA strain was subjected to IgG-Sepharose chromatography. Fractions from the final wash and MgCl2 gradient were separated by SDS-PAGE. Molecular mass standards are indicated to the left in kilodaltons. The gel was stained with Coomassie blue R.
Figure 4
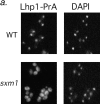
Deletion of SXM1 leads to mislocalization of Lhp1-PrA but not other nuclear proteins. (a) Wild-type haploid cells (top row) and sxm1 cells (bottom row), both with a genomic Lhp1-PrA fusion, are probed for Lhp1-PrA (left) and stained for DNA with DAPI (right). (b) Wild-type cells (top row) and sxm1 cells (bottom row) were probed for either an NLS-GFP reporter (left) or the endogenous protein Npl3p (right).
Figure 4
Deletion of SXM1 leads to mislocalization of Lhp1-PrA but not other nuclear proteins. (a) Wild-type haploid cells (top row) and sxm1 cells (bottom row), both with a genomic Lhp1-PrA fusion, are probed for Lhp1-PrA (left) and stained for DNA with DAPI (right). (b) Wild-type cells (top row) and sxm1 cells (bottom row) were probed for either an NLS-GFP reporter (left) or the endogenous protein Npl3p (right).
Figure 6
Sxm1–PrA binds to several nucleoporins but not to Kap60p. (a) Cytosol from either Kap95–PrA or Sxm1–PrA was overlaid on a nitrocellulose strip of electrophoretically separated crude E. coli lysate expressing repeat regions of Nup159p (amino acids 441–876; Kraemer et al., 1995) or Nup1p (amino acids 432– 816; Rexach and Blobel, 1995) or full-length Nsp1p or Nup2p. A strip stained for total protein with amido black is included for reference. (b) Purified Kap60p was overlaid with Kap95-PrA cytosol or Sxm1–PrA cytosol.
Similar articles
- Factors affecting nuclear export of the 60S ribosomal subunit in vivo.
Stage-Zimmermann T, Schmidt U, Silver PA. Stage-Zimmermann T, et al. Mol Biol Cell. 2000 Nov;11(11):3777-89. doi: 10.1091/mbc.11.11.3777. Mol Biol Cell. 2000. PMID: 11071906 Free PMC article. - Identification and functional characterization of a novel nuclear localization signal present in the yeast Nab2 poly(A)+ RNA binding protein.
Truant R, Fridell RA, Benson RE, Bogerd H, Cullen BR. Truant R, et al. Mol Cell Biol. 1998 Mar;18(3):1449-58. doi: 10.1128/MCB.18.3.1449. Mol Cell Biol. 1998. PMID: 9488461 Free PMC article. - Phosphorylation of the Saccharomyces cerevisiae La protein does not appear to be required for its functions in tRNA maturation and nascent RNA stabilization.
Long KS, Cedervall T, Walch-Solimena C, Noe DA, Huddleston MJ, Annan RS, Wolin SL. Long KS, et al. RNA. 2001 Nov;7(11):1589-602. RNA. 2001. PMID: 11720288 Free PMC article. - Review: transport of tRNA out of the nucleus-direct channeling to the ribosome?
Grosshans H, Simos G, Hurt E. Grosshans H, et al. J Struct Biol. 2000 Apr;129(2-3):288-94. doi: 10.1006/jsbi.2000.4226. J Struct Biol. 2000. PMID: 10806079 Review. - tRNA dynamics between the nucleus, cytoplasm and mitochondrial surface: Location, location, location.
Chatterjee K, Nostramo RT, Wan Y, Hopper AK. Chatterjee K, et al. Biochim Biophys Acta Gene Regul Mech. 2018 Apr;1861(4):373-386. doi: 10.1016/j.bbagrm.2017.11.007. Epub 2017 Nov 28. Biochim Biophys Acta Gene Regul Mech. 2018. PMID: 29191733 Free PMC article. Review.
Cited by
- Intersection of the Kap123p-mediated nuclear import and ribosome export pathways.
Sydorskyy Y, Dilworth DJ, Yi EC, Goodlett DR, Wozniak RW, Aitchison JD. Sydorskyy Y, et al. Mol Cell Biol. 2003 Mar;23(6):2042-54. doi: 10.1128/MCB.23.6.2042-2054.2003. Mol Cell Biol. 2003. PMID: 12612077 Free PMC article. - Nuclear import of histone H2A and H2B is mediated by a network of karyopherins.
Mosammaparast N, Jackson KR, Guo Y, Brame CJ, Shabanowitz J, Hunt DF, Pemberton LF. Mosammaparast N, et al. J Cell Biol. 2001 Apr 16;153(2):251-62. doi: 10.1083/jcb.153.2.251. J Cell Biol. 2001. PMID: 11309407 Free PMC article. - The Ty1 integrase protein can exploit the classical nuclear protein import machinery for entry into the nucleus.
McLane LM, Pulliam KF, Devine SE, Corbett AH. McLane LM, et al. Nucleic Acids Res. 2008 Aug;36(13):4317-26. doi: 10.1093/nar/gkn383. Epub 2008 Jun 27. Nucleic Acids Res. 2008. PMID: 18586821 Free PMC article. - Yeast Los1p has properties of an exportin-like nucleocytoplasmic transport factor for tRNA.
Hellmuth K, Lau DM, Bischoff FR, Künzler M, Hurt E, Simos G. Hellmuth K, et al. Mol Cell Biol. 1998 Nov;18(11):6374-86. doi: 10.1128/MCB.18.11.6374. Mol Cell Biol. 1998. PMID: 9774653 Free PMC article. - The yeast nuclear pore complex and transport through it.
Aitchison JD, Rout MP. Aitchison JD, et al. Genetics. 2012 Mar;190(3):855-83. doi: 10.1534/genetics.111.127803. Genetics. 2012. PMID: 22419078 Free PMC article.
References
- Aitchison JD, Blobel G, Rout MP. Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science. 1996;274:624–627. - PubMed
- Bachmann M, Pfeifer K, Schroder HC, Muller WE. Characterization of the autoantigen La as a nucleic acid-dependent ATPase/dATPase with melting properties. Cell. 1990;60:85–93. - PubMed
- Dingwall C, Laskey RA. Nuclear targeting sequences—a consensus? . Trends Biol Sci. 1991;16:478–481. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases