Aggregation as a determinant of protein fate in post-Golgi compartments: role of the luminal domain of furin in lysosomal targeting - PubMed (original) (raw)
Aggregation as a determinant of protein fate in post-Golgi compartments: role of the luminal domain of furin in lysosomal targeting
N Wolins et al. J Cell Biol. 1997.
Abstract
The mammalian endopeptidase furin is a type 1 integral membrane protein that is predominantly localized to the TGN and is degraded in lysosomes with a t1/2 = 2-4 h. Whereas the localization of furin to the TGN is largely mediated by sorting signals in the cytosolic tail of the protein, we show here that targeting of furin to lysosomes is a function of the luminal domain of the protein. Inhibition of lysosomal degradation results in the accumulation of high molecular weight aggregates of furin; aggregation is also dependent on the luminal domain of furin. Temperature and pharmacologic manipulations suggest that furin aggregation occurs in the TGN and thus precedes delivery to lysosomes. These findings are consistent with a model in which furin becomes progressively aggregated in the TGN, an event that leads to its transport to lysosomes. Our observations indicate that changes in the aggregation state of luminal domains can be potent determinants of biosynthetic targeting to lysosomes and suggest the possible existence of quality control mechanisms for disposal of aggregated proteins in compartments of the secretory pathway other than the endoplasmic reticulum.
Figures
Figure 1
Schematic representation of constructs used in this study and summary of results. (A) The diagrams show the position and number of amino acid residues of the signal peptide (SP), pro-region (PR), luminal (LU), transmembrane (TM), and cytosolic (CY) domains of furin and Tac. The figure also indicates the approximate positions of sites of N-glycan addition (N-CHO) in both molecules. The gap in the furin luminal domain represents a segment that was omitted to draw the schemes to scale. (B) The diagrams represent the structures of different furin-Tac chimeras after cleavage of the signal peptide and pro-region segments. In naming the chimeras, F and T were used to designate domains derived from furin and Tac, respectively. Some of the constructs were tagged by addition of an HA epitope (YPYDVPDYA) at the COOH terminus, as shown in the figure. In some constructs, a FLAG epitope (DYKDDDDK) was used in place of the HA epitope. The number of residues in the transmembrane and cytosolic domains of the chimeras are indicated.
Figure 2
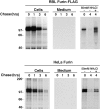
Lysosomal turnover of furin in transfected cells. Stably transfected RBL cells expressing furin-FLAG and transiently transfected HeLa cells expressing untagged furin were analyzed by pulse chase metabolic labeling as described in Materials and Methods. Cells were pulse labeled for 30 min with [35S]methionine and chased for various times in the absence (−) or presence (+) of 50 mM NH4Cl as indicated in the figure. Furin-FLAG was isolated from cells and media by immunoprecipitation with a mixture of antibodies, including Fur1, Fur2, DC16, and M2; untagged furin was isolated using the same antibody mixture, except for M2. The presence of an ∼80-kD soluble furin species in the media was detected upon prolonged exposure of the autoradiograms.
Figure 3
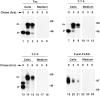
Rapid turnover of furin is not mediated by the transmembrane and cytosolic domains. Stably transfected RBL cells expressing Tac, T-T-F, T-F-F, or furin-FLAG (see Fig. 1 for structures) were pulse labeled with [35S]methionine for 30 min and chased for 0, 3, or 6 h. Tac species were isolated by immunoprecipitation with the monoclonal antibody 7G7 and furin species with a mixture of Fur1, Fur2, and DC16. The symbols p and m point to the ER precursor and Golgi-processed, mature forms of the Tac proteins, respectively. The positions of molecular weight markers (expressed as 10−3 × M r) are shown at right.
Figure 4
The luminal domain of furin is necessary and sufficient for its rapid lysosomal degradation. Stably transfected RBL cells expressing various furin and Tac chimeras (see Fig. 1 for structures) were pulse labeled with [35S]methionine for 30 min and chased for different times in the absence (−) or presence (+) of 50 mM NH4Cl. Tac and furin species were isolated with the same antibodies mentioned in the legend to Fig. 3. The positions of relative molecular mass markers (expressed as 10−3 × M r) are shown at right. The symbols p and m point to the ER precursor and Golgi-processed, mature forms of the Tac proteins, respectively. The symbols mb and s mark the positions of membrane-bound (∼100 kD) and soluble (∼80 kD) forms of furin, respectively, as previously described (Bosshart et al., 1994).
Figure 5
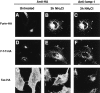
The luminal domain of furin mediates targeting to vesicles containing the lysosomal membrane protein lamp-1. Transiently transfected HeLa cells expressing furin-HA, Tac-HA, or F-T-T-HA were incubated for 3 h in the absence or presence of 50 mM NH4Cl. Cells were fixed, permeabilized, and then stained with anti-HA and anti–lamp-1 antibodies, and a mixture of tetramethyl rhodamine anti–mouse IgG and fluorescein anti–rabbit IgG. Cells were examined by confocal microscopy as described in Materials and Methods. Notice the localization of furin-HA and F-T-T-HA to vesicles containing lamp-1 in NH4Cl-treated cells (arrowheads in B, C, E, and F).
Figure 6
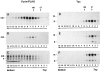
Aggregation of furin-FLAG in NH4Cl-treated cells. Stably transfected RBL cells expressing furin-FLAG, Tac, or Tac-DKQTLL were pulse labeled for 30 min with [35S]methionine and chased for 0 h (A, D, and G), 4 h (B), or 6 h (C, E, F, H, and I) in the absence (A, B, D, E, G, and H) or presence (C, F, and I) of 50 mM NH4Cl. Detergent lysates of the cells were fractionated by sedimentation on 5–20% sucrose gradients for 16 h (see Materials and Methods). Individual gradient fractions (1– 15) were immunoprecipitated with either an antifurin antibody (Fur2) (A–C) or an antibody to Tac (7G7) (D–I), and then analyzed by SDS-PAGE and fluorography. The MHC class I H-2Kb/β2-microglobulin complex (∼57 kD, fraction 3) and the transferrin receptor (∼190 kD, fractions 5 and 6) were used as internal size standards for integral membrane proteins. Gradient fraction numbers, from top to bottom, are indicated. The bracket in C indicates high molecular weight aggregates of furin-FLAG.
Figure 6
Aggregation of furin-FLAG in NH4Cl-treated cells. Stably transfected RBL cells expressing furin-FLAG, Tac, or Tac-DKQTLL were pulse labeled for 30 min with [35S]methionine and chased for 0 h (A, D, and G), 4 h (B), or 6 h (C, E, F, H, and I) in the absence (A, B, D, E, G, and H) or presence (C, F, and I) of 50 mM NH4Cl. Detergent lysates of the cells were fractionated by sedimentation on 5–20% sucrose gradients for 16 h (see Materials and Methods). Individual gradient fractions (1– 15) were immunoprecipitated with either an antifurin antibody (Fur2) (A–C) or an antibody to Tac (7G7) (D–I), and then analyzed by SDS-PAGE and fluorography. The MHC class I H-2Kb/β2-microglobulin complex (∼57 kD, fraction 3) and the transferrin receptor (∼190 kD, fractions 5 and 6) were used as internal size standards for integral membrane proteins. Gradient fraction numbers, from top to bottom, are indicated. The bracket in C indicates high molecular weight aggregates of furin-FLAG.
Figure 7
Analysis of the aggregation state of furin by chemical cross-linking. RBL cells expressing either furin-HA (A) or furin-FLAG (B) were pulse labeled with [35S]methionine for 30 min at 37°C. Cells were either lysed immediately or chased for 4 h at 37°C in the presence of 50 mM NH4Cl. Lysates were cross-linked with either 0.1 mM (A) or varying concentrations (B) of DTSSP. Furin-HA (A) was isolated by immunoprecipitation with an antibody mixture containing HA-11, Fur1, and Fur2. Furin-FLAG (B) was immunoprecipitated with M2, Fur1, and Fur2. Samples were resolved by SDS-PAGE under nonreducing or reducing conditions, as indicated in the figure. Arrows point to the position of the furin dimer. The positions of molecular mass markers (expressed as 10−3 × M r) are shown at right.
Figure 7
Analysis of the aggregation state of furin by chemical cross-linking. RBL cells expressing either furin-HA (A) or furin-FLAG (B) were pulse labeled with [35S]methionine for 30 min at 37°C. Cells were either lysed immediately or chased for 4 h at 37°C in the presence of 50 mM NH4Cl. Lysates were cross-linked with either 0.1 mM (A) or varying concentrations (B) of DTSSP. Furin-HA (A) was isolated by immunoprecipitation with an antibody mixture containing HA-11, Fur1, and Fur2. Furin-FLAG (B) was immunoprecipitated with M2, Fur1, and Fur2. Samples were resolved by SDS-PAGE under nonreducing or reducing conditions, as indicated in the figure. Arrows point to the position of the furin dimer. The positions of molecular mass markers (expressed as 10−3 × M r) are shown at right.
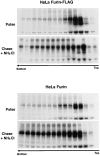
Figure 8
Both furin-FLAG and untagged furin aggregate in HeLa cells. HeLa cells were transiently transfected with plasmids encoding either furin-FLAG or untagged furin. Cells were labeled for 30 min with [35S]methionine and chased for 6 h in the presence or absence of 50 mM NH4Cl. The aggregation state of furin was assessed by sedimentation velocity analysis as described in the legend to Fig. 6.
Figure 9
The luminal domain mediates aggregation of furin. Stably transfected RBL cells expressing the furin or Tac chimeras indicated in the figure were pulse labeled for 30 min with [35S]methionine and chased for 4 h in the presence of a mixture of nonacidotropic lysosomal inhibitors (LPEM). Detergent-solubilized cells were fractionated by sedimentation on sucrose gradients and Tac and furin species were immunoprecipitated from the fractions as described in the legend to Fig. 6 and in Materials and Methods. Notice that only proteins having the furin luminal domain underwent aggregation during chase in the presence of lysosomal inhibitors.
Figure 10
Effects of AlF4 − on aggregation and degradation of furin accumulated in the TGN. (A) RBL cells expressing furin-FLAG were pulse labeled with [35S]methionine for 30 min and chased for either 0 or 6 h at 19°C. Furin-FLAG was isolated by immunoprecipitation with a mixture of the antibodies M2, Fur1, and Fur2. Immunoprecipitates were resolved by SDS-PAGE. (B) RBL cells expressing furin-FLAG were pulse labeled with [35S]methionine for 30 min and chased for 6 h at 19°C. Cells were then further chased for 30 min at 19°C and 6 h at 37°C in the absence (Untreated) or presence of aluminum fluoride (AlF4−). Cell lysates were cross-linked with 1 mM DTSSP and furin-FLAG was immunoprecipitated and analyzed as described in A.
Similar articles
- The cytoplasmic domain mediates localization of furin to the trans-Golgi network en route to the endosomal/lysosomal system.
Bosshart H, Humphrey J, Deignan E, Davidson J, Drazba J, Yuan L, Oorschot V, Peters PJ, Bonifacino JS. Bosshart H, et al. J Cell Biol. 1994 Sep;126(5):1157-72. doi: 10.1083/jcb.126.5.1157. J Cell Biol. 1994. PMID: 7914893 Free PMC article. - An acidic sequence within the cytoplasmic domain of furin functions as a determinant of trans-Golgi network localization and internalization from the cell surface.
Voorhees P, Deignan E, van Donselaar E, Humphrey J, Marks MS, Peters PJ, Bonifacino JS. Voorhees P, et al. EMBO J. 1995 Oct 16;14(20):4961-75. doi: 10.1002/j.1460-2075.1995.tb00179.x. EMBO J. 1995. PMID: 7588625 Free PMC article. - Two independent targeting signals in the cytoplasmic domain determine trans-Golgi network localization and endosomal trafficking of the proprotein convertase furin.
Schäfer W, Stroh A, Berghöfer S, Seiler J, Vey M, Kruse ML, Kern HF, Klenk HD, Garten W. Schäfer W, et al. EMBO J. 1995 Jun 1;14(11):2424-35. doi: 10.1002/j.1460-2075.1995.tb07240.x. EMBO J. 1995. PMID: 7781597 Free PMC article. - Bi-cycling the furin pathway: from TGN localization to pathogen activation and embryogenesis.
Molloy SS, Anderson ED, Jean F, Thomas G. Molloy SS, et al. Trends Cell Biol. 1999 Jan;9(1):28-35. doi: 10.1016/s0962-8924(98)01382-8. Trends Cell Biol. 1999. PMID: 10087614 Review. - Structure and function of eukaryotic proprotein processing enzymes of the subtilisin family of serine proteases.
Van de Ven WJ, Roebroek AJ, Van Duijnhoven HL. Van de Ven WJ, et al. Crit Rev Oncog. 1993;4(2):115-36. Crit Rev Oncog. 1993. PMID: 8420571 Review.
Cited by
- Diacylglycerol enrichment of endoplasmic reticulum or lipid droplets recruits perilipin 3/TIP47 during lipid storage and mobilization.
Skinner JR, Shew TM, Schwartz DM, Tzekov A, Lepus CM, Abumrad NA, Wolins NE. Skinner JR, et al. J Biol Chem. 2009 Nov 6;284(45):30941-8. doi: 10.1074/jbc.M109.013995. Epub 2009 Sep 11. J Biol Chem. 2009. PMID: 19748893 Free PMC article. - Protein quality control in the secretory pathway.
Sun Z, Brodsky JL. Sun Z, et al. J Cell Biol. 2019 Oct 7;218(10):3171-3187. doi: 10.1083/jcb.201906047. Epub 2019 Sep 19. J Cell Biol. 2019. PMID: 31537714 Free PMC article. Review. - Human herpesvirus 7 U21 tetramerizes to associate with class I major histocompatibility complex molecules.
May NA, Wang Q, Balbo A, Konrad SL, Buchli R, Hildebrand WH, Schuck P, Hudson AW. May NA, et al. J Virol. 2014 Mar;88(6):3298-308. doi: 10.1128/JVI.02639-13. Epub 2014 Jan 3. J Virol. 2014. PMID: 24390327 Free PMC article. - Functional Rescue of F508del-CFTR Using Small Molecule Correctors.
Molinski S, Eckford PD, Pasyk S, Ahmadi S, Chin S, Bear CE. Molinski S, et al. Front Pharmacol. 2012 Sep 26;3:160. doi: 10.3389/fphar.2012.00160. eCollection 2012. Front Pharmacol. 2012. PMID: 23055971 Free PMC article. - Isoform-dependent lysosomal degradation and internalization of apolipoprotein E requires autophagy proteins.
Fote GM, Geller NR, Efstathiou NE, Hendricks N, Vavvas DG, Reidling JC, Thompson LM, Steffan JS. Fote GM, et al. J Cell Sci. 2022 Jan 15;135(2):jcs258687. doi: 10.1242/jcs.258687. Epub 2022 Jan 25. J Cell Sci. 2022. PMID: 34982109 Free PMC article.
References
- Barriocanal JG, Bonifacino JS, Yuan L, Sandoval IV. Biosynthesis, glycosylation, movement through the Golgi system, and transport to lysosomes by an N-linked carbohydrate-independent mechanism of three lysosomal integral membrane proteins. J Biol Chem. 1986;261:16755–16763. - PubMed
- Bauerfeind R, Huttner WB. Biogenesis of constitutive secretory vesicles, secretory granules and synaptic vesicles. Curr Opin Cell Biol. 1993;5:628–635. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous