The complete set of predicted genes from Saccharomyces cerevisiae in a readily usable form - PubMed (original) (raw)
The complete set of predicted genes from Saccharomyces cerevisiae in a readily usable form
J R Hudson Jr et al. Genome Res. 1997 Dec.
Abstract
Nearly all of the open reading frames (ORFs) of the yeast Saccharomyces cerevisiae have been synthesized by PCR using a set of approximately 6000 primer pairs. Each of the forward primers has a common 22-base sequence at its 5' end, and each of the back primers has a common 20-base sequence at its 5' end. These common termini allow reamplification of the entire set of original PCR products using a single pair of longer primers-in our case, 70 bases. The resulting 70-base elements that flank each ORF can be used for rapid and efficient cloning into a linearized yeast vector that contains these same elements at its termini. This cloning by genetic recombination obviates the need for ligations or bacterial manipulations and should permit convenient global approaches to gene function that require the assay of each putative yeast gene.
Figures
Figure 1
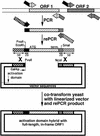
Strategy for cloning each of the yeast ORFs. A segment of the yeast genome containing illustrative ORFs 1 and 2, both of which would be transcribed left to right, is shown at the top. Each ORF is flanked by two bent arrows representing the unique pairs of primers, with the solid black fill (_rightward_-pointing arrows) indicating the common 5′ termini of the forward primers and the horizontal-lined fill (leftward pointing arrows) indicating the common 5′ termini of the back primers. The sequence of each forward primer is 5′-GGAATTCCAGCTGACCACCATGN17–29-3′ corresponding to 19 bases of non-yeast sequence, the initiator ATG of the ORF, and 17–29 subsequent bases of the ORF. The sequence of each back primer is 5′-GATCCCCGGGAATTGCCATGENDN17–29-3′ corresponding to 20 bases of non-yeast sequence, followed by the reverse complement of a stop codon (noted above as “END”), followed by the reverse complement of 17–29 bases found at the end of the ORF. The product of the PCR of ORF1, shown as a box with diagonal-lined fill flanked by the 19 and 20 bp of non-yeast sequences, is template for the rePCR. The 70-base sequences of the rePCR primers, shown as bent arrows with checkered and “brick” 5′ termini, and 3′ termini matching the sequences in the first set of PCR primers, are provided in Methods. The product of the rePCR is ORF1 flanked by 70-base elements that are identical to those in the two-hybrid vector pOAD. The positions of the translation initiation codon (ATG) and termination codon (term.) of the ORF1 rePCR product are shown. Digestion of the vector with _Nco_I and _Pvu_II and cotransformation with the rePCR product results in two recombination events that precisely insert the ORF in-frame with the activation domain of Gal4p.
Figure 2
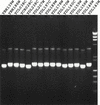
Products of a sample of rePCRs. Fifteen ORFs were reamplified using the 70-base activation domain rePCR primers. The sizes of these ORFs from the genome sequence are YBR139W (1527 bp), YBR186w (1611 bp), YCL038c (1587 bp), YCR028c (1539 bp), YDL238c (1470 bp), YDL207w (1617 bp), YDL197c (1578 bp), YDL189w (1612 bp), YDL178w (1593 bp), YDL170w (1587 bp), YDL160c (1521 bp), YDL159w (1548 bp), YDL156w (1569 bp), YDL146w (1476 bp), and YDL143w (1587 bp). The DNA size marker in the right lane has fragments of 23, 9.4, 6.6, 4.4, 2.3, 2.0, 1.4, 1.1, 0.9, and 0.6 kb. The rePCR products all migrate slightly slower than the predicted sizes of their ORFs because of the 70 bp of extra sequence at each end.
Similar articles
- The DNA sequence of cosmid 14-13b from chromosome XIV of Saccharomyces cerevisiae reveals an unusually high number of overlapping open reading frames.
De Antoni A, D'Angelo M, Dal Pero F, Sartorello F, Pandolfo D, Pallavicini A, Lanfranchi G, Valle G. De Antoni A, et al. Yeast. 1997 Mar 15;13(3):261-6. doi: 10.1002/(SICI)1097-0061(19970315)13:3<261::AID-YEA64>3.0.CO;2-L. Yeast. 1997. PMID: 9090055 - High-throughput cloning of human liver complete open reading frames using homologous recombination in Escherichia coli.
Zhu D, Zhong X, Tan R, Chen L, Huang G, Li J, Sun X, Xu L, Chen J, Ou Y, Zhang T, Yuan D, Zhang Z, Shu W, Ma L. Zhu D, et al. Anal Biochem. 2010 Feb 15;397(2):162-7. doi: 10.1016/j.ab.2009.10.018. Epub 2009 Oct 14. Anal Biochem. 2010. PMID: 19835833 - Functional analysis of the yeast genome.
Winzeler EA, Davis RW. Winzeler EA, et al. Curr Opin Genet Dev. 1997 Dec;7(6):771-6. doi: 10.1016/s0959-437x(97)80039-1. Curr Opin Genet Dev. 1997. PMID: 9468786 Review. - Unified inventory of established and putative transporters encoded within the complete genome of Saccharomyces cerevisiae.
Paulsen IT, Sliwinski MK, Nelissen B, Goffeau A, Saier MH Jr. Paulsen IT, et al. FEBS Lett. 1998 Jun 23;430(1-2):116-25. doi: 10.1016/s0014-5793(98)00629-2. FEBS Lett. 1998. PMID: 9678606 Review.
Cited by
- Structural basis for the multi-activity factor Rad5 in replication stress tolerance.
Shen M, Dhingra N, Wang Q, Cheng C, Zhu S, Tian X, Yu J, Gong X, Li X, Zhang H, Xu X, Zhai L, Xie M, Gao Y, Deng H, He Y, Niu H, Zhao X, Xiang S. Shen M, et al. Nat Commun. 2021 Jan 12;12(1):321. doi: 10.1038/s41467-020-20538-w. Nat Commun. 2021. PMID: 33436623 Free PMC article. - DNA binding and condensation properties of the herpes simplex virus type 1 triplex protein VP19C.
Bera A, Perkins EM, Zhu J, Zhu H, Desai P. Bera A, et al. PLoS One. 2014 Aug 14;9(8):e104640. doi: 10.1371/journal.pone.0104640. eCollection 2014. PLoS One. 2014. PMID: 25121591 Free PMC article. - The fate of linear DNA in Saccharomyces cerevisiae and Candida glabrata: the role of homologous and non-homologous end joining.
Corrigan MW, Kerwin-Iosue CL, Kuczmarski AS, Amin KB, Wykoff DD. Corrigan MW, et al. PLoS One. 2013 Jul 24;8(7):e69628. doi: 10.1371/journal.pone.0069628. Print 2013. PLoS One. 2013. PMID: 23894512 Free PMC article. - Preparation of recombinant protein spotted arrays for proteome-wide identification of kinase targets.
Im H, Snyder M. Im H, et al. Curr Protoc Protein Sci. 2013 Apr;Chapter 27:Unit 27.4. doi: 10.1002/0471140864.ps2704s72. Curr Protoc Protein Sci. 2013. PMID: 23546622 Free PMC article. - High-throughput cloning and expression library creation for functional proteomics.
Festa F, Steel J, Bian X, Labaer J. Festa F, et al. Proteomics. 2013 May;13(9):1381-99. doi: 10.1002/pmic.201200456. Epub 2013 Apr 5. Proteomics. 2013. PMID: 23457047 Free PMC article. Review.
References
- Bartel PL, Roecklein JA, SenGupta D, Fields S. A protein linkage map of Escherichia coli bacteriophage T7. Nature Genet. 1996;12:72–77. - PubMed
- Breeden L, Nasmyth K. Regulation of the yeast HO gene. Cold Spring Harbor Symp Quant Biol. 1985;50:643–650. - PubMed
- DeRisi J, Penland L, Brown PO, Bittner ML, Meltzer PS, Ray M, Chen Y, Su YA, Trent JM. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nature Genet. 1996;14:457–460. - PubMed
- Dujon B. The yeast genome project: What did we learn? Trends Genet. 1996;12:263–270. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases