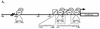The t(8;21) fusion product, AML-1-ETO, associates with C/EBP-alpha, inhibits C/EBP-alpha-dependent transcription, and blocks granulocytic differentiation - PubMed (original) (raw)
The t(8;21) fusion product, AML-1-ETO, associates with C/EBP-alpha, inhibits C/EBP-alpha-dependent transcription, and blocks granulocytic differentiation
J J Westendorf et al. Mol Cell Biol. 1998 Jan.
Abstract
AML-1B is a hematopoietic transcription factor that is functionally inactivated by multiple chromosomal translocations in human acute myeloblastic and B-cell lymphocytic leukemias. The t(8;21)(q22;q22) translocation replaces the C terminus, including the transactivation domain of AML-1B, with ETO, a nuclear protein of unknown function. We previously showed that AML-1-ETO is a dominant inhibitor of AML-1B-dependent transcriptional activation. Here we demonstrate that AML-1-ETO also inhibits C/EBP-alpha-dependent activation of the myeloid cell-specific, rat defensin NP-3 promoter. AML-1B bound the core enhancer motifs present in the NP-3 promoter and activated transcription approximately sixfold. Similarly, C/EBP-alpha bound NP-3 promoter sequences and activated transcription approximately sixfold. Coexpression of C/EBP-alpha with AML-1B or its family members, AML-2 and murine AML-3, synergistically activated the NP-3 promoter up to 60-fold. The t(8;21) product, AML-1-ETO, repressed AML-1B-dependent activation of NP-3 and completely inhibited C/EBP-alpha-dependent activity as well as the synergistic activation. In contrast, the inv(16) product, which indirectly targets AML family members by fusing their heterodimeric DNA binding partner, CBF-beta, to the myosin heavy chain, inhibited AML-1B but not C/EBP-alpha activation or the synergistic activation. AML-1-ETO and C/EBP-alpha were coimmunoprecipitated and thus physically interact in vivo. Deletion mutants demonstrated that the C terminus of ETO was required for AML-1-ETO-mediated repression of the synergistic activation but not for association with C/EBP-alpha. Finally, overexpression of AML-1-ETO in myeloid progenitor cells prevented granulocyte colony-stimulating factor-induced differentiation. Thus, AML-1-ETO may contribute to leukemogenesis by specifically inhibiting C/EBP-alpha- and AML-1B-dependent activation of myeloid promoters and blocking differentiation.
Figures
FIG. 1
AML-1B, AML-2, AML-3, and C/EBP-α bind to NP-3 promoter sequences. (A) Schematic of the rat NP-3 promoter showing locations of core binding motifs (underlined), C/EBP binding sites, and reporter constructs used in this study. (B to D) Binding of AML family members to NP-3 promoter sequences determined by electrophoretic mobility shift assays. A 32P-labeled annealed oligonucleotide probe containing the consensus core site, TGTGGT, was incubated with 3 μg of lysates from COS-7 cells overexpressing AML-1B (B), AML-2 (C), or mAML-3 (D) in the presence or absence of the indicated unlabeled competitor. The NP-3 promoter competitors were generated by PCR. (E and F) Binding of C/EBP-α to NP-3 promoter sequences. NP-3 promoter sequences extending −137 (E) and −87 (F) nucleotides from the transcription start site were labeled with [α-32P]dATP and incubated with 3 μg of C/EBP-α-overexpressing COS-7 cell lysates in the presence or absence of an unlabeled oligonucleotide containing the consensus C/EBP binding site. Supershift assays were performed with 1 μg of C/EBP-α antiserum. The location of the C/EBP-α–DNA complex is denoted by the arrows, and the supershifted complex is adjacent to the asterisks.
FIG. 1
AML-1B, AML-2, AML-3, and C/EBP-α bind to NP-3 promoter sequences. (A) Schematic of the rat NP-3 promoter showing locations of core binding motifs (underlined), C/EBP binding sites, and reporter constructs used in this study. (B to D) Binding of AML family members to NP-3 promoter sequences determined by electrophoretic mobility shift assays. A 32P-labeled annealed oligonucleotide probe containing the consensus core site, TGTGGT, was incubated with 3 μg of lysates from COS-7 cells overexpressing AML-1B (B), AML-2 (C), or mAML-3 (D) in the presence or absence of the indicated unlabeled competitor. The NP-3 promoter competitors were generated by PCR. (E and F) Binding of C/EBP-α to NP-3 promoter sequences. NP-3 promoter sequences extending −137 (E) and −87 (F) nucleotides from the transcription start site were labeled with [α-32P]dATP and incubated with 3 μg of C/EBP-α-overexpressing COS-7 cell lysates in the presence or absence of an unlabeled oligonucleotide containing the consensus C/EBP binding site. Supershift assays were performed with 1 μg of C/EBP-α antiserum. The location of the C/EBP-α–DNA complex is denoted by the arrows, and the supershifted complex is adjacent to the asterisks.
FIG. 2
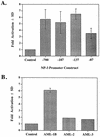
Activation of the NP-3 promoter by AML (CBF) transcription factors. (A) C33A cells were transfected with 2 to 5 μg of the indicated NP-3-Luc reporter construct, either 5 μg of RSV-SEAP or 0.5 μg of RSV-CAT as an internal control, and 1 μg of pCMV5 or pCMV5-AML-1B expression plasmid. RLU were normalized with respect to CAT or SEAP activity. Fold activation represents the normalized promoter activity from cells transfected with AML-1B relative to activity from cells transfected with NP-3-Luc alone. The basal activity was normalized to 1 for each promoter construct. (B) Effects of 1 μg of pCMV5-AML-1B, -AML-2, and -mAML-3 on NP(−137) activity were determined as described for panel A. Results represent the mean ± SD for triplicate experiments.
FIG. 3
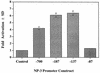
C/EBP-α activates the NP-3 promoter. C33A cells were transfected with 2 to 5 μg of the indicated NP-3-Luc reporter construct, either 5 μg of RSV-SEAP or 0.5 μg of RSV-CAT as an internal control, and 0.5 μg of pMSV or pMSV-C/EBP-α expression plasmid. RLU were normalized with respect to CAT or SEAP activity. Fold activation represents the normalized promoter activity from cells transfected with C/EBP-α relative to activity from cells transfected with NP-3-Luc alone. The basal activity was normalized to 1 for each promoter construct. Results represent the mean ± SD for triplicate experiments.
FIG. 4
C/EBP-α and AML family members cooperatively activate the NP-3 promoter. C33A cells were transfected with 5 μg of NP-3(−137); 0.5 μg of RSV-CAT; 1 μg of pCMV5 or pCMV5-AML-1B, -AML-2, or -mAML-3; and 0.5 μg of pMSV (open bars) or MSV-C/EBP-α (shaded bars). RLU were normalized with respect to CAT activity. Results represent the mean ± SD for triplicate experiments.
FIG. 5
Effects of AML-1B C-terminal deletion mutants on C/EBP-α synergism. (A) Schematic of AML1, AML-1B, and AML-1B mutants used in this study. (B to D) Synergistic effects of C/EBP-α and AML-1B or AML-1B C-terminal deletion mutants (B and C), C/EBP-α and AML-1 (D), or C/EBP-α and an AML-1B mutant lacking the ERK phosphorylation sites (E) on NP-3(−137) activity determined in C33A cells as described in the legend to Fig. 4. Fold synergy is the quotient of the actual synergistic activation and the expected additive response. Results represent the mean ± SD for triplicate experiments.
FIG. 5
Effects of AML-1B C-terminal deletion mutants on C/EBP-α synergism. (A) Schematic of AML1, AML-1B, and AML-1B mutants used in this study. (B to D) Synergistic effects of C/EBP-α and AML-1B or AML-1B C-terminal deletion mutants (B and C), C/EBP-α and AML-1 (D), or C/EBP-α and an AML-1B mutant lacking the ERK phosphorylation sites (E) on NP-3(−137) activity determined in C33A cells as described in the legend to Fig. 4. Fold synergy is the quotient of the actual synergistic activation and the expected additive response. Results represent the mean ± SD for triplicate experiments.
FIG. 6
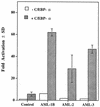
Effects of AML-1–ETO, AML-1–ETO truncation mutants, and inv(16) on C/EBP-α- and AML-1B-induced synergistic activation of the NP-3 promoter. (A) Schematic of the AML-1–ETO and inv(16) proteins used in this study. (B to D) Transfection of C33A cells with 5 μg of NP-3(−137), 0.5 μg of RSV-CAT, 1 μg of pCMV5 or CMV5-AML-1B expression plasmid, 0.5 μg of pMSV or MSV-C/EBP-α expression plasmid, or both CMV5-AML-1B and MSV-C/EBP-α in the presence of 10 μg of pCMV5 (control) or pCMV5-AML-1–ETO (B), pCMV5-AML-1–ETO-L148D (C), or pCMV5-inv(16) (D). RLU were normalized with respect to CAT activity. Results represent the mean ± SD for triplicate experiments.
FIG. 6
Effects of AML-1–ETO, AML-1–ETO truncation mutants, and inv(16) on C/EBP-α- and AML-1B-induced synergistic activation of the NP-3 promoter. (A) Schematic of the AML-1–ETO and inv(16) proteins used in this study. (B to D) Transfection of C33A cells with 5 μg of NP-3(−137), 0.5 μg of RSV-CAT, 1 μg of pCMV5 or CMV5-AML-1B expression plasmid, 0.5 μg of pMSV or MSV-C/EBP-α expression plasmid, or both CMV5-AML-1B and MSV-C/EBP-α in the presence of 10 μg of pCMV5 (control) or pCMV5-AML-1–ETO (B), pCMV5-AML-1–ETO-L148D (C), or pCMV5-inv(16) (D). RLU were normalized with respect to CAT activity. Results represent the mean ± SD for triplicate experiments.
FIG. 7
C/EBP-α physically associates with AML-1–ETO but not ETO in vivo. COS-7 cells were transfected with the indicated pCMV5 expression plasmid(s) and metabolically labeled for 3 h with [35S]methionine. Lysates were immunoprecipitated with antiserum to the N terminus of AML1 (A), C/EBP-α (B and D), or ETO (C) and analyzed by denaturing sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis. Numbers at the left indicate the migration of molecular weight standards.
FIG. 8
ETO sequences are required for C/EBP-α inhibition by AML-1–ETO. C33A cells were transfected with 2.5 μg of the indicated pCMV5-AML-1–ETO construct, 5 μg of NP-3(−137), 0.5 μg of RSV-CAT, 1 μg of pCMV5 or CMV5-AML-1B expression plasmid, and 0.5 μg of pMSV or MSV-C/EBP-α expression plasmid. Data were corrected as described in the legend to Fig. 6. Results represent the mean ± SD for triplicate experiments.
FIG. 9
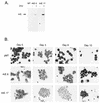
The t(8;21) fusion protein blocks granulocyte differentiation. (A) AML-1–ETO expression in 32D.3 clones containing empty vector (lane 1, MT) or expressing AML-1–ETO (lanes 2 to 5, clones 6 and 17) before and after 6 h of incubation in 400 μM zinc sulfate was determined by immunoblot analysis with ETO-specific antisera. (B) G-418-resistant control (pMT) and two clonal cell lines expressing AML-1–ETO (A/E) were cultured in the presence of G-CSF. At the indicated times, cells were cytocentrifuged and Wright stained. Note that at day 6, the control but not the cells expressing AML-1–ETO contained multilobed nuclei, which are characteristic of granulocyte maturation. By day 12, few viable cells remained in the control cell cultures, whereas cells expressing the fusion protein maintained an immature morphology.
Similar articles
- AML-2 is a potential target for transcriptional regulation by the t(8;21) and t(12;21) fusion proteins in acute leukemia.
Meyers S, Lenny N, Sun W, Hiebert SW. Meyers S, et al. Oncogene. 1996 Jul 18;13(2):303-12. Oncogene. 1996. PMID: 8710369 - The MYND motif is required for repression of basal transcription from the multidrug resistance 1 promoter by the t(8;21) fusion protein.
Lutterbach B, Sun D, Schuetz J, Hiebert SW. Lutterbach B, et al. Mol Cell Biol. 1998 Jun;18(6):3604-11. doi: 10.1128/MCB.18.6.3604. Mol Cell Biol. 1998. PMID: 9584201 Free PMC article. - Specific protein redirection as a transcriptional therapy approach for t(8;21) leukemia.
Steffen B, Serve H, Berdel WE, Agrawal S, Linggi B, Büchner T, Hiebert SW, Müller-Tidow C. Steffen B, et al. Proc Natl Acad Sci U S A. 2003 Jul 8;100(14):8448-53. doi: 10.1073/pnas.1330293100. Epub 2003 Jun 20. Proc Natl Acad Sci U S A. 2003. PMID: 12819347 Free PMC article. - Mechanisms of transcriptional repression by the t(8;21)-, t(12;21)-, and inv(16)-encoded fusion proteins.
Heibert SW, Lutterbach B, Durst K, Wang L, Linggi B, Wu S, Wood L, Amann J, King D, Hou Y. Heibert SW, et al. Cancer Chemother Pharmacol. 2001 Aug;48 Suppl 1:S31-4. doi: 10.1007/s002800100302. Cancer Chemother Pharmacol. 2001. PMID: 11587363 Review. - The AML1 gene: a transcription factor involved in the pathogenesis of myeloid and lymphoid leukemias.
Lo Coco F, Pisegna S, Diverio D. Lo Coco F, et al. Haematologica. 1997 May-Jun;82(3):364-70. Haematologica. 1997. PMID: 9234595 Review.
Cited by
- Mutational cooperativity of RUNX1::RUNX1T1 isoform 9a and oncogenic NRAS in zebrafish myeloid leukaemia.
Lints R, Walker CA, Delfi O, Prouse M, PohLui De Silva M, Bohlander SK, Wood AC. Lints R, et al. Biol Open. 2024 Sep 15;13(9):bio060523. doi: 10.1242/bio.060523. Epub 2024 Aug 30. Biol Open. 2024. PMID: 39177514 Free PMC article. - Restoration of MYC-repressed targets mediates the negative effects of GM-CSF on RUNX1-ETO leukemogenicity.
Weng S, Matsuura S, Mowery CT, Stoner SA, Lam K, Ran D, Davis AG, Lo MC, Zhang DE. Weng S, et al. Leukemia. 2017 Jan;31(1):159-169. doi: 10.1038/leu.2016.167. Epub 2016 Jun 15. Leukemia. 2017. PMID: 27389055 Free PMC article. - Hematopoietic stem cell expansion and distinct myeloid developmental abnormalities in a murine model of the AML1-ETO translocation.
de Guzman CG, Warren AJ, Zhang Z, Gartland L, Erickson P, Drabkin H, Hiebert SW, Klug CA. de Guzman CG, et al. Mol Cell Biol. 2002 Aug;22(15):5506-17. doi: 10.1128/MCB.22.15.5506-5517.2002. Mol Cell Biol. 2002. PMID: 12101243 Free PMC article. - Tribbles homolog 2 inactivates C/EBPalpha and causes acute myelogenous leukemia.
Keeshan K, He Y, Wouters BJ, Shestova O, Xu L, Sai H, Rodriguez CG, Maillard I, Tobias JW, Valk P, Carroll M, Aster JC, Delwel R, Pear WS. Keeshan K, et al. Cancer Cell. 2006 Nov;10(5):401-11. doi: 10.1016/j.ccr.2006.09.012. Cancer Cell. 2006. PMID: 17097562 Free PMC article. - Loss of C/EBP alpha cell cycle control increases myeloid progenitor proliferation and transforms the neutrophil granulocyte lineage.
Porse BT, Bryder D, Theilgaard-Mönch K, Hasemann MS, Anderson K, Damgaard I, Jacobsen SE, Nerlov C. Porse BT, et al. J Exp Med. 2005 Jul 4;202(1):85-96. doi: 10.1084/jem.20050067. Epub 2005 Jun 27. J Exp Med. 2005. PMID: 15983063 Free PMC article.
References
- Bae S C, Takahashi E, Zhang Y W, Ogawa E, Shigesada K, Namba Y, Satake M, Ito Y. Cloning, mapping and expression of PEBP2 alpha C, a third gene encoding the mammalian Runt domain. Gene. 1995;159:245–248. - PubMed
- Banaiee, N., et al. Unpublished data.
- Birkenmeier E H, Gwynn B, Howard S, Jerry J, Gordon J I, Landschulz W H, McKnight S L. Tissue-specific expression, developmental regulation, and genetic mapping of the gene encoding CCAAT/enhancer binding protein. Genes Dev. 1989;3:1146–1156. - PubMed
- Cameron S, Taylor D S, TePas E C, Speck N A, Mathey-Prevot B. Identification of a critical regulatory site in the human interleukin-3 promoter by in vivo footprinting. Blood. 1994;83:2851–2859. - PubMed
- Cao Z, Umek R M, McKnight S L. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev. 1991;5:1538–1552. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA077274/CA/NCI NIH HHS/United States
- CA64140/CA/NCI NIH HHS/United States
- R01 CA064140/CA/NCI NIH HHS/United States
- CA77274/CA/NCI NIH HHS/United States
- CA77176/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous