Twenty proteins containing a C-terminal SOCS box form five structural classes - PubMed (original) (raw)
Twenty proteins containing a C-terminal SOCS box form five structural classes
D J Hilton et al. Proc Natl Acad Sci U S A. 1998.
Abstract
The four members of the recently identified suppressor of cytokines signaling family (SOCS-1, SOCS-2, SOCS-3, and CIS, where CIS is cytokine-inducible SH2-containing protein) appear, by various means, to negatively regulate cytokine signal transduction. Structurally, the SOCS proteins are composed of an N-terminal region of variable length and amino acid composition, a central SH2 domain, and a previously unrecognized C-terminal motif that we have called the SOCS box. By using the SOCS box amino acid sequence consensus, we have searched DNA databases and have identified a further 16 proteins that contain this motif. These proteins fall into five classes based on the protein motifs found N-terminal of the SOCS box. In addition to four new SOCS proteins (SOCS-4 to SOCS-7) containing an SH2 domain and a SOCS box, we describe three new families of proteins that contain either WD-40 repeats (WSB-1 and -2), SPRY domains (SSB-1 to -3) or ankyrin repeats (ASB-1 to -3) N-terminal of the SOCS box. In addition, we show that a class of small GTPases also contains a SOCS box. The expression of representative members of each class of proteins differs markedly, as does the regulation of expression by cytokines. The function of the WSB, SSB, and ASB protein families remains to be determined.
Figures
Figure 1
Comparison of the amino acid sequence of the SOCS proteins. (A) Schematic representation of the structures of SOCS, WSB, SSB, ASB, and GTPase proteins. Green bars indicate SH2 domains, blue bars indicate WD-40 repeats, pink bars indicate ankyrin repeats, apricot bars indicate SPRY domains, magenta bars indicate GTPase domains, and yellow bars represent the SOCS box. The shaded bars represent additional less well conserved parts of the proteins. (B) Alignment of N-terminal regions of SOCS, WSB, SSB, and ASB proteins. (C) Alignment of the SH2 domains of CIS and SOCS-1 to SOCS-7. (D) Alignment of the WD-40 repeats of WSB-1, 2. (E) Alignment of the SPRY domains of SSB-1 to SSB-3. (F) Alignment of the ankyrin repeats of ASB-1 to ASB-3. (G) Alignment of the regions between SH2, WD-40, or ankyrin repeats and the SOCS box. (H) Alignment of the SOCS box of the SOCS, WSB, SSB, ASB, and GTPases. X, residues of uncertain identity; •, beginning and end of contigs. Amino acid sequence obtained by conceptual translation of ESTs is approximate only. Residues conserved among 75% or more of the proteins in a given alignment, defined as LIVMA, FYW, DE, QN, CST, KRH, and PG are shaded green in the SH2 domain, blue in the WD-40 repeats, pink in the ankyrin repeats, apricot in the SPRY domain, yellow in the SOCS box, and grey elsewhere in the protein. Alignment of SH2 domains, WD-40 repeats, SPRY domains, and ankyrin repeats has been carried out with reference to previous descriptions of these protein motifs (–17, 19). The sequences for human SOCS-6 and SOCS-7 were obtained from the following GenBank entries describing CIS4 and and Nck/Ash binding protein (GenBank accession nos. AB006968 and AB005216).
Figure 1
Comparison of the amino acid sequence of the SOCS proteins. (A) Schematic representation of the structures of SOCS, WSB, SSB, ASB, and GTPase proteins. Green bars indicate SH2 domains, blue bars indicate WD-40 repeats, pink bars indicate ankyrin repeats, apricot bars indicate SPRY domains, magenta bars indicate GTPase domains, and yellow bars represent the SOCS box. The shaded bars represent additional less well conserved parts of the proteins. (B) Alignment of N-terminal regions of SOCS, WSB, SSB, and ASB proteins. (C) Alignment of the SH2 domains of CIS and SOCS-1 to SOCS-7. (D) Alignment of the WD-40 repeats of WSB-1, 2. (E) Alignment of the SPRY domains of SSB-1 to SSB-3. (F) Alignment of the ankyrin repeats of ASB-1 to ASB-3. (G) Alignment of the regions between SH2, WD-40, or ankyrin repeats and the SOCS box. (H) Alignment of the SOCS box of the SOCS, WSB, SSB, ASB, and GTPases. X, residues of uncertain identity; •, beginning and end of contigs. Amino acid sequence obtained by conceptual translation of ESTs is approximate only. Residues conserved among 75% or more of the proteins in a given alignment, defined as LIVMA, FYW, DE, QN, CST, KRH, and PG are shaded green in the SH2 domain, blue in the WD-40 repeats, pink in the ankyrin repeats, apricot in the SPRY domain, yellow in the SOCS box, and grey elsewhere in the protein. Alignment of SH2 domains, WD-40 repeats, SPRY domains, and ankyrin repeats has been carried out with reference to previous descriptions of these protein motifs (–17, 19). The sequences for human SOCS-6 and SOCS-7 were obtained from the following GenBank entries describing CIS4 and and Nck/Ash binding protein (GenBank accession nos. AB006968 and AB005216).
Figure 2
Northern blot analysis of mRNA expression of mouse SOCS-1, SOCS-5, WSB-2, and ASB-1 in normal adult mouse tissues and in mouse embryos. Northern blot analysis of poly(A)+ mRNA extracted from a range of adult mouse tissues (Left) and mouse embryos and yolk sac (Right) as described (4). Sal. Gland, salivary gland; MLN, mesenteric lymph node; E9–E18, embryos from the 9th through to the 18th day of gestation; P1, 1 day after birth. Size markers in kilobases (kb) are shown to the left of the figure, and the identity of probes is shown to the right.
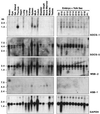
Figure 3
Northern blot analysis of mRNA expression of mouse SOCS-1, SOCS-5, WSB-2, and ASB-1 in the mouse liver before and various times after injection of IL-6. Northern blot analysis of poly(A)+ mRNA expression in the liver of mice injected with saline (sal) or IL-6. Mice were injected intravenously with 200 μl of IL-6 (25 μg/ml) or saline. At the times indicated, mice were killed, their livers were removed, and mRNA was extracted as described (4). Size markers in kilobases (kb) are shown to the left of the figure and the identity of probes is shown to the right.
Similar articles
- Suppressors of cytokine signalling: SOCS.
Larsen L, Röpke C. Larsen L, et al. APMIS. 2002 Dec;110(12):833-44. doi: 10.1034/j.1600-0463.2002.1101201.x. APMIS. 2002. PMID: 12645661 Review. - A family of cytokine-inducible inhibitors of signalling.
Starr R, Willson TA, Viney EM, Murray LJ, Rayner JR, Jenkins BJ, Gonda TJ, Alexander WS, Metcalf D, Nicola NA, Hilton DJ. Starr R, et al. Nature. 1997 Jun 26;387(6636):917-21. doi: 10.1038/43206. Nature. 1997. PMID: 9202125 - Identification of critical residues required for suppressor of cytokine signaling-specific regulation of interleukin-4 signaling.
Haque SJ, Harbor PC, Williams BR. Haque SJ, et al. J Biol Chem. 2000 Aug 25;275(34):26500-6. doi: 10.1074/jbc.275.34.26500. J Biol Chem. 2000. PMID: 10950967 - Mutational analyses of the SOCS proteins suggest a dual domain requirement but distinct mechanisms for inhibition of LIF and IL-6 signal transduction.
Nicholson SE, Willson TA, Farley A, Starr R, Zhang JG, Baca M, Alexander WS, Metcalf D, Hilton DJ, Nicola NA. Nicholson SE, et al. EMBO J. 1999 Jan 15;18(2):375-85. doi: 10.1093/emboj/18.2.375. EMBO J. 1999. PMID: 9889194 Free PMC article. - The CIS family: negative regulators of JAK-STAT signaling.
Yoshimura A. Yoshimura A. Cytokine Growth Factor Rev. 1998 Sep-Dec;9(3-4):197-204. doi: 10.1016/s1359-6101(98)00019-7. Cytokine Growth Factor Rev. 1998. PMID: 9918119 Review.
Cited by
- Functional complementation of two splicing variants of Gustavus in Neocaridina denticulata sinensis during ovarian maturation.
Liang M, Feng D, Zhang J, Sun Y. Liang M, et al. Sci Rep. 2024 Sep 9;14(1):20939. doi: 10.1038/s41598-024-72080-0. Sci Rep. 2024. PMID: 39251721 Free PMC article. - Are peptidomimetics the compounds of choice for developing new modulators of the JAK-STAT pathway?
Cugudda A, La Manna S, Marasco D. Cugudda A, et al. Front Immunol. 2024 Jun 24;15:1406886. doi: 10.3389/fimmu.2024.1406886. eCollection 2024. Front Immunol. 2024. PMID: 38983855 Free PMC article. Review. - SOCS1 is a critical checkpoint in immune homeostasis, inflammation and tumor immunity.
Bidgood GM, Keating N, Doggett K, Nicholson SE. Bidgood GM, et al. Front Immunol. 2024 Jun 14;15:1419951. doi: 10.3389/fimmu.2024.1419951. eCollection 2024. Front Immunol. 2024. PMID: 38947335 Free PMC article. Review. - Peripheral Neuropathy and Decreased Locomotion of a RAB40B Mutation in Human and Model Animals.
Son W, Jeong HS, Nam DE, Lee AJ, Nam SH, Lee JE, Choi BO, Chung KW. Son W, et al. Exp Neurobiol. 2023 Dec 31;32(6):410-422. doi: 10.5607/en23027. Exp Neurobiol. 2023. PMID: 38196136 Free PMC article. - The complementary roles of STAT3 and STAT1 in cancer biology: insights into tumor pathogenesis and therapeutic strategies.
Wang W, Lopez McDonald MC, Kim C, Ma M, Pan ZT, Kaufmann C, Frank DA. Wang W, et al. Front Immunol. 2023 Nov 8;14:1265818. doi: 10.3389/fimmu.2023.1265818. eCollection 2023. Front Immunol. 2023. PMID: 38022653 Free PMC article. Review.
References
- Nicola N A, editor. Guidebook to Cytokines and Their Receptors. Oxford: Oxford Univ. Press; 1994.
- Darnell J E, Jr, Kerr I M, Stark G R. Science. 1994;264:1415–1421. - PubMed
- Ihle J N, Kerr I M. Trends Genet. 1995;11:69–74. - PubMed
- Starr R, Willson T A, Viney E M, Murray L J, Rayner J R, Jenkins B J, Gonda T J, Alexander W S, Metcalf D, Nicola N A, Hilton D J. Nature (London) 1997;387:917–921. - PubMed
- Endo T A, Masuhara M, Yokouchi M, Suzuki R, Mitsui K, Sakamoto H, Ohtsubo M, Misawa H, Kanekura Y, Yoshimura A. Nature (London) 1997;387:921–924. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases