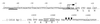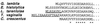A mitochondrial-like chaperonin 60 gene in Giardia lamblia: evidence that diplomonads once harbored an endosymbiont related to the progenitor of mitochondria - PubMed (original) (raw)
A mitochondrial-like chaperonin 60 gene in Giardia lamblia: evidence that diplomonads once harbored an endosymbiont related to the progenitor of mitochondria
A J Roger et al. Proc Natl Acad Sci U S A. 1998.
Abstract
Diplomonads, parabasalids, as represented by trichomonads, and microsporidia are three protist lineages lacking mitochondria that branch earlier than all other eukaryotes in small subunit rRNA and elongation factor phylogenies. The absence of mitochondria and plastids in these organisms suggested that they diverged before the origin of these organelles. However, recent discoveries of mitochondrial-like heat shock protein 70 and/or chaperonin 60 (cpn60) genes in trichomonads and microsporidia imply that the ancestors of these two groups once harbored mitochondria or their endosymbiotic progenitors. In this report, we describe a mitochondrial-like cpn60 homolog from the diplomonad parasite Giardia lamblia. Northern and Western blots reveal that the expression of cpn60 is independent of cellular stress and, except during excystation, occurs throughout the G. lamblia life cycle. Phylogenetic analyses position the G. lamblia cpn60 in a clade that includes mitochondrial and hydrogenosomal cpn60 proteins. The most parsimonious interpretation of these data is that the cpn60 gene was transferred from the endosymbiotic ancestors of mitochondria to the nucleus early in eukaryotic evolution, before the divergence of the diplomonads and trichomonads from other extant eukaryotic lineages. A more complicated explanation requires that these genes originated from distinct alpha-proteobacterial endosymbioses that formed transiently within these protist lineages.
Figures
Figure 1
Properties of the G. lamblia cpn60 gene and mRNA transcript. Upstream and downstream regions of the cpn60 gene are shown and numbered relative to the first base of the start codon (+1). The transcriptional start sites are shown in bold under vertical lines and rightward-pointing arrow (→). Two possible transcriptional signals are identified: an AT-rich transcription initiation signal (single underline), and an upstream promoter element (double underline) similar to the CAATTT signal reported for other G. lamblia genes (33). Possible sites of polyadenylation (↓), a putative polyadenylation signal (boxed), and the stop codon (∗) are also indicated.
Figure 2
Expression of cpn60 at different stages in the G. lamblia life cycle. (A) Northern blot analysis of total cellular RNA using a PCR fragment of the G lamblia cpn60 as a probe. Expression of the ≈1.8-kb cpn60 transcript was monitored in the vegetative trophozoite (T) stage, and after 5, 24, and 48 hr of encystation of G. lamblia. (B) Western blot analysis of total cellular protein isolated at different stages of encystation and excysation using a Synechococcus sp. anti-cpn60 antibody. Levels of the ≈60-kDa putative cpn60 protein were constant during the transition from the trophozoite stage (T) and throughout 0–66 hr of encystation. This protein was also present in the cyst phase (C) and after the first stage of induction of excystation (pI). A marked decrease in cpn60 protein was observed after the second stage of encystation (pII). Low levels of cpn60 persisted at 20 min, 90 min, 1 day, and 3 days after excystation, increasing after 6 days.
Figure 3
An alignment of the N termini of G. lamblia, E. histolytica, mitochondrial, hydrogenosomal, and eubacterial cpn60 homologs. The deduced N-terminal sequence of G. lamblia and E. histolytica cpn60s are aligned with homologs from L. tarentolae, T. vaginalis, and Caulobacter crescentus. The L. tarentolae and T. vaginalis targeting peptides (underlined) are removed during import into mitochondria and hydrogenosomes, respectively (9, 39). An N-terminal extension of the E. histolytica cpn60 homolog is suggestive of a targeting peptide for a cryptic organelle in this organism. G. lamblia cpn60 has a small, 2-aa N-terminal extension relative to C. crescentus. Amino acid identities of the G. lamblia cpn60 to other homologs are indicated by asterisks (∗) under the alignment.
Figure 4
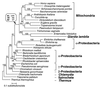
Phylogenetic relationships of cpn60 homologs. Protein maximum likelihood analysis of 513 aligned amino acid positions yielded the tree shown (log likelihood = −18676.09). Optimal trees obtained by using protein distance and maximum parsimony methods differed from this topology in the branching order of the major eukaryotic groups and, to a lesser extent, within the α-, β-, and γ-proteobacterial clades. The branching order between these clades was identical for all methods. Bootstrap values obtained for the _G. lamblia/_mitochondrial-like cpn60 clade when using protein maximum likelihood (ML), protein distance matrix (DM), and maximum parsimony (MP) methods are shown in a box above the relevant node (indicated by an arrow). For all other nodes in the tree, protein ML bootstrap values (where >50%) are shown above each branch. The scale bar indicates estimated sequence divergence per unit branch length.
Similar articles
- A possible mitochondrial gene in the early-branching amitochondriate protist Trichomonas vaginalis.
Roger AJ, Clark CG, Doolittle WF. Roger AJ, et al. Proc Natl Acad Sci U S A. 1996 Dec 10;93(25):14618-22. doi: 10.1073/pnas.93.25.14618. Proc Natl Acad Sci U S A. 1996. PMID: 8962102 Free PMC article. - Primary structure and phylogenetic relationships of a malate dehydrogenase gene from Giardia lamblia.
Roger AJ, Morrison HG, Sogin ML. Roger AJ, et al. J Mol Evol. 1999 Jun;48(6):750-5. doi: 10.1007/pl00006519. J Mol Evol. 1999. PMID: 10229579 - Phylogenetic affinity of a Giardia lamblia cysteine desulfurase conforms to canonical pattern of mitochondrial ancestry.
Emelyanov VV. Emelyanov VV. FEMS Microbiol Lett. 2003 Sep 26;226(2):257-66. doi: 10.1016/S0378-1097(03)00598-6. FEMS Microbiol Lett. 2003. PMID: 14553920 - Rickettsiaceae, rickettsia-like endosymbionts, and the origin of mitochondria.
Emelyanov VV. Emelyanov VV. Biosci Rep. 2001 Feb;21(1):1-17. doi: 10.1023/a:1010409415723. Biosci Rep. 2001. PMID: 11508688 Review. - Evolution of the chaperonin families (Hsp60, Hsp10 and Tcp-1) of proteins and the origin of eukaryotic cells.
Gupta RS. Gupta RS. Mol Microbiol. 1995 Jan;15(1):1-11. doi: 10.1111/j.1365-2958.1995.tb02216.x. Mol Microbiol. 1995. PMID: 7752884 Review.
Cited by
- On the origin of the nucleus: a hypothesis.
Baum B, Spang A. Baum B, et al. Microbiol Mol Biol Rev. 2023 Dec 20;87(4):e0018621. doi: 10.1128/mmbr.00186-21. Epub 2023 Nov 29. Microbiol Mol Biol Rev. 2023. PMID: 38018971 Free PMC article. Review. - Unusual Cell Structures and Organelles in Giardia intestinalis and Trichomonas vaginalis Are Potential Drug Targets.
Benchimol M, Gadelha AP, de Souza W. Benchimol M, et al. Microorganisms. 2022 Nov 2;10(11):2176. doi: 10.3390/microorganisms10112176. Microorganisms. 2022. PMID: 36363768 Free PMC article. Review. - Import of Entamoeba histolytica Mitosomal ATP Sulfurylase Relies on Internal Targeting Sequences.
Santos HJ, Chiba Y, Makiuchi T, Arakawa S, Murakami Y, Tomii K, Imai K, Nozaki T. Santos HJ, et al. Microorganisms. 2020 Aug 12;8(8):1229. doi: 10.3390/microorganisms8081229. Microorganisms. 2020. PMID: 32806678 Free PMC article. - Prenylquinones in Human Parasitic Protozoa: Biosynthesis, Physiological Functions, and Potential as Chemotherapeutic Targets.
Verdaguer IB, Zafra CA, Crispim M, Sussmann RAC, Kimura EA, Katzin AM. Verdaguer IB, et al. Molecules. 2019 Oct 16;24(20):3721. doi: 10.3390/molecules24203721. Molecules. 2019. PMID: 31623105 Free PMC article. Review. - Five facts about Giardia lamblia.
Cernikova L, Faso C, Hehl AB. Cernikova L, et al. PLoS Pathog. 2018 Sep 27;14(9):e1007250. doi: 10.1371/journal.ppat.1007250. eCollection 2018 Sep. PLoS Pathog. 2018. PMID: 30261050 Free PMC article. Review. No abstract available.
References
- Leipe D D, Gunderson J H, Nerad T A, Sogin M L. Mol Biochem Parasitol. 1993;59:41–48. - PubMed
- Sogin M L, Gunderson J H, Elwood H J, Alonso R A, Peattie D A. Science. 1989;243:75–77. - PubMed
- Hashimoto T, Hasegawa M. Adv Biophys. 1996;32:73–120. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM53835/GM/NIGMS NIH HHS/United States
- AI24285/AI/NIAID NIH HHS/United States
- P01 DK035108/DK/NIDDK NIH HHS/United States
- DK35108/DK/NIDDK NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Research Materials