Phylogeny of the genus Flavivirus - PubMed (original) (raw)
Comparative Study
Phylogeny of the genus Flavivirus
G Kuno et al. J Virol. 1998 Jan.
Abstract
We undertook a comprehensive phylogenetic study to establish the genetic relationship among the viruses of the genus Flavivirus and to compare the classification based on molecular phylogeny with the existing serologic method. By using a combination of quantitative definitions (bootstrap support level and the pairwise nucleotide sequence identity), the viruses could be classified into clusters, clades, and species. Our phylogenetic study revealed for the first time that from the putative ancestor two branches, non-vector and vector-borne virus clusters, evolved and from the latter cluster emerged tick-borne and mosquito-borne virus clusters. Provided that the theory of arthropod association being an acquired trait was correct, pairwise nucleotide sequence identity among these three clusters provided supporting data for a possibility that the non-vector cluster evolved first, followed by the separation of tick-borne and mosquito-borne virus clusters in that order. Clades established in our study correlated significantly with existing antigenic complexes. We also resolved many of the past taxonomic problems by establishing phylogenetic relationships of the antigenically unclassified viruses with the well-established viruses and by identifying synonymous viruses.
Figures
FIG. 1
Relative genomic positions of primers used for amplification of DNA templates from the NS5 gene of flaviviruses and for sequencing. ∗, From reference . ∗∗, The name in parentheses indicates a degenerate primer at the same location corresponding to the primer immediately above. ∗∗∗, From reference .
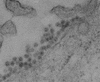
FIG. 2
Electron micrograph of Vero cells infected with Tamana bat virus (magnification, × 121,125).
FIG. 3
Phylogenetic tree of the genus Flavivirus, using nucleotide sequence. The tree was constructed by the neighbor-joining method of MEGA. Each number at nodes is the percentage of 500 bootstrap replicate support; ∗ indicates confidence probability higher than 90%. Vertical length is arbitrary. Scale is percentage of genetic distance.
FIG. 4
Phylogenetic tree of the genus Flavivirus, using amino acid sequence. The tree was constructed by the neighbor-joining method using MEGA. Each number at nodes is the percentage of 500 bootstrap replicates. Vertical length is arbitrary. Scale is percentage of amino acid distance.
FIG. 5
Multiple amino acid sequence alignment of the CFA, Apoi, and Kedougou viruses and one member representing each clade of the genus Flavivirus. Amino acid 1 corresponds to nucleotides 9018 to 9020 of YF virus. A dash indicates missing one amino acid. A dot indicates that the amino acid (or absence of it) in a given amino acid sequence is the same as in the corresponding sequence of CFA virus above of the aligned sequences.
FIG. 6
Pairwise nucleotide sequence identity relationship among three clusters of the genus Flavivirus. ∗, Number of pairs with 63 to 65% nucleotide sequence identity/total number of pair sequence compared.
Similar articles
- Phylogenetic relationships of flaviviruses correlate with their epidemiology, disease association and biogeography.
Gaunt MW, Sall AA, Lamballerie X, Falconar AKI, Dzhivanian TI, Gould EA. Gaunt MW, et al. J Gen Virol. 2001 Aug;82(Pt 8):1867-1876. doi: 10.1099/0022-1317-82-8-1867. J Gen Virol. 2001. PMID: 11457992 - Complete genomic sequence of Powassan virus: evaluation of genetic elements in tick-borne versus mosquito-borne flaviviruses.
Mandl CW, Holzmann H, Kunz C, Heinz FX. Mandl CW, et al. Virology. 1993 May;194(1):173-84. doi: 10.1006/viro.1993.1247. Virology. 1993. PMID: 8097605 - Phylogeny of TYU, SRE, and CFA virus: different evolutionary rates in the genus Flavivirus.
Marin MS, Zanotto PM, Gritsun TS, Gould EA. Marin MS, et al. Virology. 1995 Feb 1;206(2):1133-9. doi: 10.1006/viro.1995.1038. Virology. 1995. PMID: 7856087 - Origins, evolution, and vector/host coadaptations within the genus Flavivirus.
Gould EA, de Lamballerie X, Zanotto PM, Holmes EC. Gould EA, et al. Adv Virus Res. 2003;59:277-314. doi: 10.1016/s0065-3527(03)59008-x. Adv Virus Res. 2003. PMID: 14696332 Review. - Mechanisms Underlying Host Range Variation in Flavivirus: From Empirical Knowledge to Predictive Models.
Halabi K, Mayrose I. Halabi K, et al. J Mol Evol. 2021 Jul;89(6):329-340. doi: 10.1007/s00239-021-10013-5. Epub 2021 May 31. J Mol Evol. 2021. PMID: 34059925 Review.
Cited by
- Development and characterization of specific anti-Usutu virus chicken-derived single chain variable fragment antibodies.
Schoenenwald AKJ, Gwee CP, Stiasny K, Hermann M, Vasudevan SG, Skern T. Schoenenwald AKJ, et al. Protein Sci. 2020 Nov;29(11):2175-2188. doi: 10.1002/pro.3937. Epub 2020 Sep 4. Protein Sci. 2020. PMID: 32829514 Free PMC article. - Isolation of Infective Zika Virus from Urine and Saliva of Patients in Brazil.
Bonaldo MC, Ribeiro IP, Lima NS, Dos Santos AA, Menezes LS, da Cruz SO, de Mello IS, Furtado ND, de Moura EE, Damasceno L, da Silva KA, de Castro MG, Gerber AL, de Almeida LG, Lourenço-de-Oliveira R, Vasconcelos AT, Brasil P. Bonaldo MC, et al. PLoS Negl Trop Dis. 2016 Jun 24;10(6):e0004816. doi: 10.1371/journal.pntd.0004816. eCollection 2016 Jun. PLoS Negl Trop Dis. 2016. PMID: 27341420 Free PMC article. - New oxindole carboxamides as inhibitors of DENV NS5 RdRp: Design, synthesis, docking and Biochemical characterization.
Koraboina CP, Akshinthala P, Katari NK, Adarasandi R, Jonnalagadda SB, Gundla R. Koraboina CP, et al. Heliyon. 2023 Nov 2;9(11):e21510. doi: 10.1016/j.heliyon.2023.e21510. eCollection 2023 Nov. Heliyon. 2023. PMID: 38027588 Free PMC article. - Zika virus NS1 structure reveals diversity of electrostatic surfaces among flaviviruses.
Song H, Qi J, Haywood J, Shi Y, Gao GF. Song H, et al. Nat Struct Mol Biol. 2016 May;23(5):456-8. doi: 10.1038/nsmb.3213. Epub 2016 Apr 18. Nat Struct Mol Biol. 2016. PMID: 27088990 - Evolutionary relationships of endemic/epidemic and sylvatic dengue viruses.
Wang E, Ni H, Xu R, Barrett AD, Watowich SJ, Gubler DJ, Weaver SC. Wang E, et al. J Virol. 2000 Apr;74(7):3227-34. doi: 10.1128/jvi.74.7.3227-3234.2000. J Virol. 2000. PMID: 10708439 Free PMC article.
References
- Ando K, Kuratsuka K, Arima S, Hironaka N, Honda Y, Ishii K. Studies on the viruses isolated during epidemic of Japanese B encephalitis in 1948 in Tokyo area. Kitasato Arch Exp Med. 1952;24:49–61. - PubMed
- Berthet, F.-X., H. G. Zeller, M.-T. Drouet, J. Rauzier, J.-P. Digoutte, and V. Deubel. Extensive nucleotide changes and deletions within the envelope glycoprotein gene of Euro-African West Nile viruses. J. Gen. Virol. 78:2293–2297. - PubMed
- Blok J, Gibbs A J. Molecular systematics of the flaviviruses and their relatives. In: Gibbs A, Calisher C H, Garcia-Arenal F, editors. Molecular basis of virus evolution. Cambridge, England: Cambridge University Press; 1995. pp. 270–289.
- Bowen M D, Peters C J, Mills J N, Nichol S T. Oliveiros virus: a novel arenavirus from Argentina. Virology. 1996;217:362–366. - PubMed
- Calisher C H. Antigenic classification and taxonomy of flaviviruses (family Flaviviridae) emphasizing a universal system for the taxonomy of viruses causing tick-borne encephalitis. Acta Virol. 1988;32:469–478. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous