Sindbis virus induces apoptosis through a caspase-dependent, CrmA-sensitive pathway - PubMed (original) (raw)
Sindbis virus induces apoptosis through a caspase-dependent, CrmA-sensitive pathway
V E Nava et al. J Virol. 1998 Jan.
Abstract
Sindbis virus infection of cultured cells and of neurons in mouse brains leads to programmed cell death exhibiting the classical characteristics of apoptosis. Although the mechanism by which Sindbis virus activates the cell suicide program is not known, we demonstrate here that Sindbis virus activates caspases, a family of death-inducing proteases, resulting in cleavage of several cellular substrates. To study the role of caspases in virus-induced apoptosis, we determined the effects of specific caspase inhibitors on Sindbis virus-induced cell death. CrmA (a serpin from cowpox virus) and zVAD-FMK (N-benzyloxycarbonyl-Val-Ala-Asp-fluoromethyl ketone) inhibited Sindbis virus-induced cell death, suggesting that cellular caspases facilitate apoptosis induced by Sindbis virus. Furthermore, CrmA significantly increased the rate of survival of infected mice. These inhibitors appear to protect cells by inhibiting the cellular death pathway rather than impairing virus replication or by inhibiting the nsP2 and capsid viral proteases. The specificity of CrmA indicates that the Sindbis virus-induced death pathway is similar to that induced by Fas or tumor necrosis factor alpha rather than being like the death pathway induced by DNA damage. Taken together, these data suggest a central role for caspases in Sindbis virus-induced apoptosis.
Figures
FIG. 1
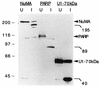
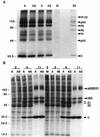
Cleavage of cellular proteins by caspases during Sindbis virus-induced apoptosis. BHK cells were mock infected or infected (MOI, 10) with wild-type Sindbis virus strain AR339. Lysates were collected at 24 h postinfection and immunoblotted with three different affinity-purified antibodies obtained from the sera of individuals with autoimmune disease. The positions of intact NuMA, PARP, and U1-70kDa proteins and their cleavage products are indicated. The results are representative of two independent experiments. The positions of molecular size markers (in kilodaltons) are shown on the left.
FIG. 2
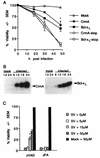
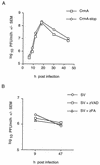
CrmA and zVAD-FMK protect BHK cells from Sindbis virus-induced cell death. (A) BHK cells were mock infected or infected with recombinant Sindbis viruses encoding the indicated genes, and cell viability was determined at the indicated times by trypan blue exclusion. Data from three to nine independent experiments are shown. Error bars (indication standard deviations) are hidden by the symbol at some time points. (B) Immunoblots of N18 cells infected with recombinant viruses encoding CrmA (left) or Bcl-xL (right). Cells were harvested at the indicated times (in hours) postinfection, and equal amounts of protein were analyzed by SDS–15% PAGE. Similar results were obtained with BHK cells (data not shown). (C) The viabilities of Sindbis virus (AR339)-infected BHK cells treated with zVAD-FMK or zFA-FMK at the indicated concentrations were determined by trypan blue exclusion at 48 h postinfection. The results summarize data from three to eight independent experiments. The asterisk indicates a statistically significant difference upon comparison of the viability of each recombinant virus with that of its corresponding stop construct in panel A and upon comparison of the viability of cells treated with 15 μM zVAD-FMK with that of the other categories in panel C (P < 0.05 by Student’s t test).
FIG. 3
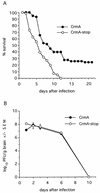
CrmA enhances survival of Sindbis virus-infected mice. (A) Percent survival of mice infected with the indicated recombinant viruses was determined in four independent experiments with approximately 40 mice (total) per virus. A P value of <0.002 was obtained by life table analysis. (B) Replication of recombinant viruses in mouse brains was determined by plaque assay. Each datum point represents the geometric mean virus titer ± the standard error of the mean (SEM) of values for three mouse brains.
FIG. 4
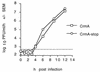
One-step growth curves of with recombinant viruses with and without CrmA show no differences. BHK cells were infected with CrmA or CrmA-stop recombinant virus (MOI, 10), and progeny viruses produced during a 1-h period were collected from the supernatants and titered by plaque assay. The function of CrmA and CrmA-stop was confirmed by determining cell viability in control wells. The results represent the means of two independent experiments plaqued in duplicate for each time point. Bars for standard error of the mean (SEM) are hidden by the symbols, and the dashed horizontal line marks the limit of detection.
FIG. 5
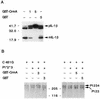
CrmA does not alter production of nonstructural and structural Sindbis virus proteins. (A) Sindbis virus nonstructural proteins (P123, P1, P2, and P3) were immunoprecipitated from equal volumes of labeled lysates, prepared 11 h after infection, with recombinant viruses encoding crmA (A), crmA-stop (AS), bcl-xL (X), and bcl-xL-stop (XS) or from mock-infected cells (M) with rabbit antiserum raised against nsP2. The nonstructural proteins were identified by their molecular masses (the positions of molecular size markers [in kilodaltons] are shown on the left), by comparison to published protein patterns (15), and by comparison to nonstructural proteins immunoprecipitated with a mixture of antibodies to nsP2 and nsP3 (X2) provided by M. Gorrell and D. Griffin. Proteins were resolved by SDS–10% PAGE and processed with salicylic acid prior to autoradiography. The results are representative of three independent experiments. (B) Sindbis virus structural proteins were analyzed by SDS–15% PAGE and autoradiography of labeled whole-cell lysates harvested at the indicated times (in hours) after infection of BHK cells. These results are representative of six independent time course experiments. The arrows indicate the precursor viral glycoproteins (pE3E2E1 and pE2), the mature glycoproteins (E1 and E2), and the capsid protein (C). Molecular mass standards (in kilodaltons) are indicated.
FIG. 6
Caspase inhibitors CrmA and zVAD-FMK do not significantly inhibit Sindbis virus replication. (A) Recombinant viruses produced in BHK cell supernatants (MOI, 10) during 1-h intervals were collected, and plaque assays were performed in duplicate. Each datum point is the mean of values for three independent wells harvested on the same day, and the results are representative of seven independent experiments. (B) Production of progeny Sindbis virus (AR339) per hour in BHK cells treated with zVAD-FMK or zFA-FMK was determined. The cells were pretreated with 50 μM zVAD-FMK or zFA-FMK for 2 h, and the inhibitors were replenished after 24 h. Each datum point is the mean of values for three independent wells harvested on the same day.
FIG. 7
CrmA inhibits caspase 1 but does not inhibit nsP2 protease activity in vitro. (A) In vitro-translated pIL-1β was digested with caspase 1 in the presence (1 or 5 μl) or absence (−) of purified GST-CrmA protein and then analyzed by SDS–10% PAGE. Cleavage of pIL-1β to the 17.5-kDa mature form (mIL-1β) was inhibited by GST-CrmA but not by GST protein. (B) Labeled in vitro-translated nsP1234 (C481G) was digested with unlabeled in vitro-translated nsP2 protease (P1*2*3) in the presence (3 or 5 μl) or absence (−) of purified GST-CrmA or GST protein alone. Labeled nsP123 and molecular mass standards (in kilodaltons) serve as markers.
FIG. 8
Model of the Sindbis virus-induced apoptotic pathway. Different death stimuli appear to activate distinct upstream caspases, as determined on the basis of inhibitor profiles. Upstream caspases activate downstream caspases, leading to cell death. The presence of an alternate caspase-independent pathway cannot be ruled out.
Similar articles
- Involvement of caspase family proteases in transforming growth factor-beta-induced apoptosis.
Chen RH, Chang TY. Chen RH, et al. Cell Growth Differ. 1997 Jul;8(7):821-7. Cell Growth Differ. 1997. PMID: 9218876 - Cytotoxic T lymphocyte-assisted suicide. Caspase 3 activation is primarily the result of the direct action of granzyme B.
Atkinson EA, Barry M, Darmon AJ, Shostak I, Turner PC, Moyer RW, Bleackley RC. Atkinson EA, et al. J Biol Chem. 1998 Aug 14;273(33):21261-6. doi: 10.1074/jbc.273.33.21261. J Biol Chem. 1998. PMID: 9694885 - Need for caspases in apoptosis of trophic factor-deprived PC12 cells.
Haviv R, Lindenboim L, Li H, Yuan J, Stein R. Haviv R, et al. J Neurosci Res. 1997 Oct 1;50(1):69-80. doi: 10.1002/(SICI)1097-4547(19971001)50:1<69::AID-JNR8>3.0.CO;2-J. J Neurosci Res. 1997. PMID: 9379495 - Regulators of apoptosis on the road to persistent alphavirus infection.
Griffin DE, Hardwick JM. Griffin DE, et al. Annu Rev Microbiol. 1997;51:565-92. doi: 10.1146/annurev.micro.51.1.565. Annu Rev Microbiol. 1997. PMID: 9343360 Review. - Neuronal apoptosis pathways in Sindbis virus encephalitis.
Irusta PM, Hardwick JM. Irusta PM, et al. Prog Mol Subcell Biol. 2004;36:71-93. doi: 10.1007/978-3-540-74264-7_5. Prog Mol Subcell Biol. 2004. PMID: 15171608 Review.
Cited by
- Inhibitor specificity of recombinant and endogenous caspase-9.
Ryan CA, Stennicke HR, Nava VE, Burch JB, Hardwick JM, Salvesen GS. Ryan CA, et al. Biochem J. 2002 Sep 1;366(Pt 2):595-601. doi: 10.1042/BJ20020863. Biochem J. 2002. PMID: 12067274 Free PMC article. - Effects of inducing or inhibiting apoptosis on Sindbis virus replication in mosquito cells.
Wang H, Blair CD, Olson KE, Clem RJ. Wang H, et al. J Gen Virol. 2008 Nov;89(Pt 11):2651-2661. doi: 10.1099/vir.0.2008/005314-0. J Gen Virol. 2008. PMID: 18931060 Free PMC article. - Recovery from viral encephalomyelitis: immune-mediated noncytolytic virus clearance from neurons.
Griffin DE. Griffin DE. Immunol Res. 2010 Jul;47(1-3):123-33. doi: 10.1007/s12026-009-8143-4. Immunol Res. 2010. PMID: 20087684 Free PMC article. Review. - The role of innate versus adaptive immune responses in a mouse model of O'nyong-nyong virus infection.
Seymour RL, Rossi SL, Bergren NA, Plante KS, Weaver SC. Seymour RL, et al. Am J Trop Med Hyg. 2013 Jun;88(6):1170-9. doi: 10.4269/ajtmh.12-0674. Epub 2013 Apr 8. Am J Trop Med Hyg. 2013. PMID: 23568285 Free PMC article. - Characterization of reemerging chikungunya virus.
Sourisseau M, Schilte C, Casartelli N, Trouillet C, Guivel-Benhassine F, Rudnicka D, Sol-Foulon N, Le Roux K, Prevost MC, Fsihi H, Frenkiel MP, Blanchet F, Afonso PV, Ceccaldi PE, Ozden S, Gessain A, Schuffenecker I, Verhasselt B, Zamborlini A, Saïb A, Rey FA, Arenzana-Seisdedos F, Desprès P, Michault A, Albert ML, Schwartz O. Sourisseau M, et al. PLoS Pathog. 2007 Jun;3(6):e89. doi: 10.1371/journal.ppat.0030089. PLoS Pathog. 2007. PMID: 17604450 Free PMC article.
References
- Ahmad M, Srinivasula S M, Wang L, Litwack G, Fernandes-Alnemri T, Alnemri E S. Spodoptera frugiperda caspase-1, a novel insect death protease that cleaves the nuclear immunophilin FKBP46, is the target of the baculovirus antiapoptotic protein p35. J Biol Chem. 1997;272:1421–1424. - PubMed
- Boldin M P, Goncharov T M, Goltsev Y V, Wallach D. Involvement of MACH, a novel MORT1/FADD-interacting protease, in Fas/APO-1- and TNF receptor-induced cell death. Cell. 1996;85:803–815. - PubMed
- Boulakia C A, Chen G, Ng F W, Teodoro J G, Branton P E, Nicholson D W, Poirier G G, Shore G C. Bcl-2 and adenovirus E1B 19 kDa protein prevent E1A-induced processing of CPP32 and cleavage of poly(ADP-ribose) polymerase. Oncogene. 1996;12:529–535. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous