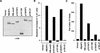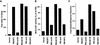Stable binding to E2F is not required for the retinoblastoma protein to activate transcription, promote differentiation, and suppress tumor cell growth - PubMed (original) (raw)
Stable binding to E2F is not required for the retinoblastoma protein to activate transcription, promote differentiation, and suppress tumor cell growth
W R Sellers et al. Genes Dev. 1998.
Abstract
The retinoblastoma tumor suppressor protein (pRB) can inhibit cell cycle progression and promote differentiation. pRB interacts with a variety of transcription factors, including members of the E2F and C-EBP protein families and MyoD, and can either repress or activate transcription depending on the promoter under study. These biological and biochemical activities of pRB have been mapped previously to a core domain, referred to as the pRB pocket. Using a panel of synthetic pRB pocket mutants, we found that the acute induction of a G1/S block by pRB is linked to its ability to both bind to E2F and to repress transcription. In contrast, these functions were not required for pRB to promote differentiation, which correlated with its ability to activate transcription in concert with fate-determining proteins such as MyoD. All tumor-derived pRB mutants tested to date failed to bind to E2F and did not repress transcription. Despite an inability to bind to E2F, pRB mutants associated with a low risk of retinoblastoma, unlike high-risk mutants, retained the ability to activate transcription and promote differentiation. Thus, the pRB pocket participates in dual tumor suppressor functions, one linked to cell cycle progression and the other to differentiation control, and these functions can be genetically and mechanistically dissociated.
Figures
Figure 1
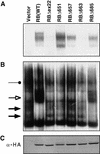
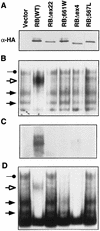
E2F-binding to pRB mutants. (A) IP–DOC release. SAOS2 cells were transfected with plasmids encoding epitope HA-tagged versions of the indicated pRB proteins. Anti-HA immunoprecipitates were prepared and bound proteins were released by treatment with deoxycholate. Released proteins were scored in gel-shift assays with a 32P-radiolabeled E2F DNA-binding site. (B) E2F DNA-binding activities in transiently transfected cells. Nuclear extracts were prepared from SAOS2 cells transiently transfected as in A and analyzed in gel-shift assays with a 32P-radiolabeled E2F DNA-binding site. (Solid arrows) Complexes containing free E2F; (open arrow and solid circles) pRB/E2F and p107/E2F complexes, respectively. (C) Production of pRB mutants. Whole cell extracts (150 μg) prepared from SAOS2 cells transiently transfected as in A were immunoblotted with an anti-HA antibody.
Figure 2
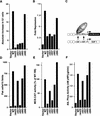
Transcriptional repression and activation functions predict the ability of pRB to induce a cell cycle block and promote differentiation. (A) Induction of a G1 arrest by pRB. SAOS2 cells were transiently transfected with plasmids encoding the indicated pRB proteins along with a plasmid encoding the cell-surface marker CD19. Seventy-two hours later, the DNA content of CD19-positive cells was determined by FACS. The _y_-axis indicates the absolute increase in the percentage of cells in G1 relative to cells transfected with the backbone expression plasmid. Each value represents the mean of 2–5 independent experiments. (B) Transcriptional repression mediated by TETr–RB(379-928) and mutant derivatives. SAOS2 cells were cotransfected with pGL2ANΔTETo, pCMV-β-Gal and plasmids encoding either wild-type TETr–RB(379-928) or the indicated mutant derivatives. Luciferase values were determined and normalized for β-galactosidase activity. Fold repression represents the corrected luciferase value obtained with TETr alone divided by the corrected luciferase obtained with the indicated TETr–RB chimeras. Data shown are means of replicate values and are representative of three independent experiments. (C) Experimental design for repression assays. Transcriptional repression was measured by use of a luciferase reporter plasmid (pGL2ANΔTETo) containing the region from −211 to +64 of the E2F1 promoter (Neuman et al. 1994). Both E2F DNA-binding sites were replaced by TETo DNA-binding sites. Wild-type pRB(379–928), and mutant derivatives thereof, were targeted to DNA as chimeras containing the DNAbinding domain of TETr (TETr–RBLP) (Sellers et al. 1995). (D) Flat cell induction. SAOS2 cells were transfected with plasmids encoding the indicated pRB proteins along with a neomycin resistance plasmid. The number of flat cells per ten 100× fields following 2 weeks of G418 selection was determined by manual counting. Values represent the mean of four independent experiments. (E) Transactivation of the MCK promoter. RB −/− MEFs were cotransfected with plasmids encoding the indicated pRB proteins along with a plasmid encoding MyoD and a plasmid containing the MCK promoter upstream of CAT. CAT activity was determined 36–48 hr following transfection. CAT activity relative to the activity obtained in the presence of wild-type RB is shown. Data are representative of two independent experiments. (F) Induction of alkaline phosphatase activity. SAOS2 cells were transfected and selected in G418 as in D. Alkaline phosphatase activity was measured using a colorimetric substrate and normalized for total protein content. Values represent the means of duplicate samples and are representative of three independent experiments.
Figure 3
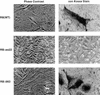
Morphological changes induced following restoration of pRB function in SAOS2 osteogenic sarcoma cells. SAOS2 osteogenic sarcoma cells were stably transfected with plasmids encoding the indicated pRB proteins. (Left) Phase-contrast micrographs of cells following 2 weeks of selection in G418. (Right) von Kossa Staining for mineral deposition after 2 weeks selection in G418 followed by 2 weeks incubation in media containing 50 μg/ml
l
-ascorbic acid and 10 m
m
β-glycerol-phosphate. Magnification, 300×.
Figure 4
An E2F1–RB chimera does not induce flat cells. SAOS2 cells were transfected with plasmids encoding either wild-type pRB, pRBΔex22, or the indicated E2F1-RB chimeras, along with a neomycin resistance marker. (E2F1) E2F1(1–368); (RBSP) pRB(379–792); (RBLP) pRB(379–928). The number of flat cells per ten 100× fields following 2 weeks of G418 selection was determined by manual counting. Data shown are representative of three independent experiments. (Top) The ability of the various proteins to either repress transcription or to induce a G1/S arrest, as determined previously (Sellers et al. 1995). (Bottom) Production of each protein was confirmed by anti-RB immunoprecipitation followed by anti-RB immunoblot analysis.
Figure 5
pRB family members differ in flat cell induction. (A) Production of pRB family members in SAOS2 cells. SAOS2 cells transiently transfected with plasmids encoding the indicated HA-tagged wild-type or mutant pRB family members were lysed and immunoprecipitated with an anti-HA antibody. The immunoprecipitates were resolved by electrophoresis in a 7.5% SDS–polyacrylamide gel and immunoblotted with an anti-HA antibody. (B) pRB family members induce a cell cycle block in SAOS2 cells. SAOS2 cells were transiently transfected with plasmids encoding the indicated wild-type or mutant pRB family members along with a plasmid encoding the cell surface marker CD19. Seventy-two hours later the DNA content of CD19-positive cells was determined by FACS. The _y_-axis indicates the absolute increase in the percentage of cells in G1 relative to cells transfected with the backbone expression plasmid. Data shown is from a single experiment and is representative of data obtained in three independent experiments. (C) Flat cell induction. SAOS2 cells were transfected with plasmids encoding the indicated wild-type or mutant pRB family members, along with a neomycin resistance plasmid. The number of flat cells per ten 100× fields following 2 weeks of G418 selection was determined by manual counting. Data shown are from a single experiment and is representative of data obtained in three independent experiments.
Figure 6
Proteins encoded by partially penetrant RB-1 alleles are defective for E2F binding. (A,C) SAOS2 cells transiently transfected with plasmids encoding the indicated HA-tagged pRB proteins were lysed and immunoprecipitated with an anti-HA antibody. The immunoprecipitates were immunoblotted with an anti-HA antibody (A) or were assayed for coimmunoprecipitated E2F by gel-shift analysis of deoxycholate released proteins by use of a 32P-radiolabeled E2F DNA-binding site (C). (B) E2F DNA-binding activity in transiently transfected cells. Nuclear extracts were prepared from SAOS2 cells transiently transfected with plasmids encoding the indicated HA-tagged pRB proteins and analyzed in gel-shift assays using an 32P-radiolabeled E2F DNA-binding site. (Solid arrows) Complexes containing free E2F; (open arrow and solid circle) pRB/E2F and p107/E2F complexes, respectively. (D) E2F complexes in stably transfected cells. SAOS2 cells were transfected with plasmids encoding the indicated HA-tagged pRB proteins along with a neomycin resistance plasmid. Following 2 weeks of selection in media containing G418, nuclear extracts were prepared and analyzed as in B.
Figure 7
Proteins encoded by partially penetrant RB-1 alleles preserve the ability to induce flat cells and to cooperate with MyoD. (A) Flat cell induction. SAOS2 cells were transfected with plasmids encoding the indicated HA-tagged pRB proteins along with a neomycin resistance plasmid. The number of flat cells per ten 100× fields was determined after G418 selection for 2 weeks. (B) Transactivation of the muscle creatine kinase promoter. RB −/− MEFs were cotransfected with plasmids encoding the indicated pRB proteins along with a plasmid encoding MyoD and a plasmid containing the MCK promoter upstream of CAT. CAT activity relative to that obtained in the presence of wild-type RB is shown. Data are representative of two independent experiments. (C) Transactivation of a glucocorticoid responsive promoter. SAOS2 cells were transiently transfected in duplicate with plasmids encoding the indicated HA-tagged pRB proteins along with a plasmid encoding GRα, an MTV-promoter luciferase reporter plasmid and a plasmid encoding β-galactosidase. Dexamethasone was added 16 hr after transfection to one of each duplicate. Fold dexamethasone induced activity after correction for β-galactosidase activity is shown. Data shown are representative of two independent experiments.
Figure 8
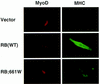
Myogenic conversion of embryonic fibroblasts. RB −/− MEFs were transiently transfected with plasmids encoding MyoD and the indicated pRB proteins. Differentiation media were added to the cells 24–48 hr after transfection. Two to four days later cells were immunostained for MyoD and myosin heavy chain (MHC) as indicated.
Figure 9
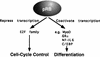
Model for pRB function.
Similar articles
- Activity of the retinoblastoma family proteins, pRB, p107, and p130, during cellular proliferation and differentiation.
Sidle A, Palaty C, Dirks P, Wiggan O, Kiess M, Gill RM, Wong AK, Hamel PA. Sidle A, et al. Crit Rev Biochem Mol Biol. 1996 Jun;31(3):237-71. doi: 10.3109/10409239609106585. Crit Rev Biochem Mol Biol. 1996. PMID: 8817077 Review. - p130, p107, and pRb are differentially regulated in proliferating cells and during cell cycle arrest by alpha-interferon.
Thomas NS, Pizzey AR, Tiwari S, Williams CD, Yang J. Thomas NS, et al. J Biol Chem. 1998 Sep 11;273(37):23659-67. doi: 10.1074/jbc.273.37.23659. J Biol Chem. 1998. PMID: 9726970 - pRB and p107/p130 are required for the regulated expression of different sets of E2F responsive genes.
Hurford RK Jr, Cobrinik D, Lee MH, Dyson N. Hurford RK Jr, et al. Genes Dev. 1997 Jun 1;11(11):1447-63. doi: 10.1101/gad.11.11.1447. Genes Dev. 1997. PMID: 9192872 - pRB, p107 and p130 as transcriptional regulators: role in cell growth and differentiation.
Mayol X, Graña X. Mayol X, et al. Prog Cell Cycle Res. 1997;3:157-69. doi: 10.1007/978-1-4615-5371-7_13. Prog Cell Cycle Res. 1997. PMID: 9580269 Review.
Cited by
- Understanding pRb: toward the necessary development of targeted treatments for retinoblastoma.
Sachdeva UM, O'Brien JM. Sachdeva UM, et al. J Clin Invest. 2012 Feb;122(2):425-34. doi: 10.1172/JCI57114. Epub 2012 Feb 1. J Clin Invest. 2012. PMID: 22293180 Free PMC article. Review. - Activation of retinoblastoma protein in mammary gland leads to ductal growth suppression, precocious differentiation, and adenocarcinoma.
Jiang Z, Zacksenhaus E. Jiang Z, et al. J Cell Biol. 2002 Jan 7;156(1):185-98. doi: 10.1083/jcb.200106084. Epub 2002 Jan 3. J Cell Biol. 2002. PMID: 11777937 Free PMC article. - Transcriptional down-regulation of the retinoblastoma protein is associated with differentiation and apoptosis in human colorectal epithelial cells.
Guy M, Moorghen M, Bond JA, Collard TJ, Paraskeva C, Williams AC. Guy M, et al. Br J Cancer. 2001 Feb;84(4):520-8. doi: 10.1054/bjoc.2000.1635. Br J Cancer. 2001. PMID: 11207048 Free PMC article. - Detection of peptides, proteins, and drugs that selectively interact with protein targets.
Serebriiskii IG, Mitina O, Pugacheva EN, Benevolenskaya E, Kotova E, Toby GG, Khazak V, Kaelin WG, Chernoff J, Golemis EA. Serebriiskii IG, et al. Genome Res. 2002 Nov;12(11):1785-91. doi: 10.1101/gr.450702. Genome Res. 2002. PMID: 12421766 Free PMC article. - Ras Regulates Rb via NORE1A.
Barnoud T, Donninger H, Clark GJ. Barnoud T, et al. J Biol Chem. 2016 Feb 5;291(6):3114-23. doi: 10.1074/jbc.M115.697557. Epub 2015 Dec 16. J Biol Chem. 2016. PMID: 26677227 Free PMC article.
References
- Adams PD, Kaelin WG. Transcriptional control by E2F. Cancer Biol. 1995;6:99–108. - PubMed
- Adnane J, Shao Z, Robbins PD. The retinoblastoma susceptibility gene product represses transcription when directly bound to the promoter. J Biol Chem. 1995;270:8837–8843. - PubMed
- Asahina I, Sampath TK, Hauschka PV. Human Osteogenic Protein-1 induces chondroblastic, osteoblastic, and/or adipocyte differentiation of clonal murine target cells. Exp Cell Res. 1996;222:38–47. - PubMed
- Baker SJ, Markowitz S, Fearon E, Willson B, Vogelstein B. Suppression of human colorectal carcinoma cell growth by wild-type p53. Science. 1990;249:912–915. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials