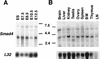The tumor suppressor gene Smad4/Dpc4 is required for gastrulation and later for anterior development of the mouse embryo - PubMed (original) (raw)
. 1998 Jan 1;12(1):107-19.
doi: 10.1101/gad.12.1.107.
J L de la Pompa, A Elia, A Itie, C Mirtsos, A Cheung, S Hahn, A Wakeham, L Schwartz, S E Kern, J Rossant, T W Mak
Affiliations
- PMID: 9420335
- PMCID: PMC316400
- DOI: 10.1101/gad.12.1.107
The tumor suppressor gene Smad4/Dpc4 is required for gastrulation and later for anterior development of the mouse embryo
C Sirard et al. Genes Dev. 1998.
Abstract
Mutations in the SMAD4/DPC4 tumor suppressor gene, a key signal transducer in most TGFbeta-related pathways, are involved in 50% of pancreatic cancers. Homozygous Smad4 mutant mice die before day 7.5 of embryogenesis. Mutant embryos have reduced size, fail to gastrulate or express a mesodermal marker, and show abnormal visceral endoderm development. Growth retardation of the Smad4-deficient embryos results from reduced cell proliferation rather than increased apoptosis. Aggregation of mutant Smad4 ES cells with wild-type tetraploid morulae rescues the gastrulation defect. These results indicate that Smad4 is initially required for the differentiation of the visceral endoderm and that the gastrulation defect in the epiblast is secondary and non-cell autonomous. Rescued embryos show severe anterior truncations, indicating a second important role for Smad4 in anterior patterning during embryogenesis.
Figures
Figure 1
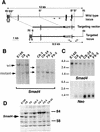
Targeted disruption of the Smad4 locus results in a null mutation. (A) Partial genomic organization of the wild-type Smad4 locus in mouse (top). The bar represents the 0.5-kb flanking probe used in Southern blot analysis generating a 12-kb _Eco_RI fragment for the wild-type allele. Arrows depict the location of the primers used in PCR analysis to identify the wild-type allele. The targeting vector (middle) was generated by cloning the 4.5-kb _Spe_I genomic fragment as the long-arm and the 385-bp _Bgl_II genomic fragment as the short-arm. The targeted locus after homologous recombination (bottom) would generate an 8.5-kb _Eco_RI fragment by use of the 0.5-kb flanking probe. Arrows depict the primers used for PCR screening of the homologous recombination in ES cells and for subsequent genotyping. An asterisk (*) indicates that the restriction site has been lost during the cloning process. (RI) _Eco_RI, (S) _Spe_I, (B) _Bgl_II. (B) Southern blot analysis of ES cell clones generated by homologous recombination at the Smad4 locus. Genomic DNA was isolated from wild type (E14K), heterozygous (C8, F9, and C8-3), and homozygous (C8-24, F9-2, and F9-5) mutant cell lines and digested with _Eco_RI. (C) Northern blot analysis of the ES cell clones. The blotted membrane containing total RNA was probed with the same 0.5-kb genomic fragment described above (Smad4) and stripped and reprobed with a neomycin specific probe (Neo). A strip and reprobe of this same membrane with the ribosomal L32 probe showed equal loading of RNA in all lanes (data not shown). (D) Western blot analysis of the ES cell clones. Total cell lysates were immunoblotted with a polyclonal α-Smad4 antibody. A band of an apparent molecular weight of 62 kd was detected in the wild-type (E14K) and the heterozygous mutants, but not in the Smad4 homozygous mutants.
Figure 2
Expression of Smad4 during embryogensis and in adult tissues. (A) Northern blot analysis of Smad4 ES cells, embryos at different developmental stages (E7.5–E12.5), and in adult tissues (B). The membranes were hybridized with the murine full length Smad4 cDNA. A prominent transcript of ∼3.8 kb was ubiquitously expressed at all developmental stages and in all adult tissues examined. Filters were stripped and reprobed with the ribosomal gene, L32, to normalize for loading variations. (BM) bone marrow, (LN) lymph node.
Figure 3
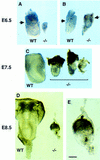
Growth retardation and poor differentiation in Smad4 mutant embryos. (A) E6.5 Smad4 mutant embryos severely growth-retarded and unorganized as compared with wild-type littermates (left); (B) mutant embryos with a poorly defined extraembryonic region. The arrows point to the separation between the embryonic and extraembryonic regions. (C) E7.5 Smad4 mutant embryos have not developed considerably and start to be resorbed. (D) E8.5 wild-type embryos start organogenesis (left). While most of the Smad4 mutant embryos are in resorption, very few remnant embryos remain with no distinguishable structures (right on D and detail in E). (Bar) 70 μm in A,B; 140 μm in C, 60 μm in D, and 20 μm in E.
Figure 4
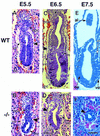
Defective mesoderm and visceral endoderm development in Smad4 mutant embryos. Embryos were sectioned and stained with hematoxylin–eosin. (A) E5.5 wild-type embryo is an early egg-cylinder stage. (B) E5.5 Smad4 mutant embryo, the embryonic region is reduced and the visceral endoderm is disorganized. (C) E6.5 wild-type late egg-cylinder stage embryo, both the extraembryonic and embryonic regions are well organized and mesoderm tissue can be distinguished. (D) E6.5 Smad4 mutant embryo, the extraembryonic region, including the visceral endoderm, is severely disorganized. The embryonic region is reduced in size and poorly developed but displays a proamniotic cavity. No sign of mesoderm can be observed. (E) E7.5 wild-type embryo, three germ layers are apparent. (F) E7.5 Smad4 mutant embryo. The arrow points to the extremely reduced embryonic region. (A_–_D) The large arrowhead points to the separation between the embryonic and extraembryonic region. (al) allantois; (ee) embryonic ectoderm; (eee) extraembryonic ectoderm; (epc) ectoplacental cone; (m) mesoderm; (pa) proamniotic cavity; (ve) visceral endoderm. (Bar) 60 μm in (A_–_D,F); 150 μm in E.
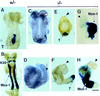
Figure 5
E6.5 Smad4 mutant embryos do not express Brachyury and show reduced Hnf4 expression and BrdU incorporation. In situ hybridization showing ubiquitous Smad4 expression in a wild-type embryo under bright-field (A) and dark-field (B) view. Smad4 expression in mutant embryo under bright-field (C) and dark-field (D) is drastically reduced. (A,C) Arrow points to the separation between the embryonic and extraembryonic regions. (E) Immunohistochemical analysis of T protein in wild-type embryo reveals T-positive cells in the nascent streak (arrowhead). (F) Absence of T expression in Smad4 mutant embryo. In situ hybridization showing Hnf4 expression in the visceral endoderm (ve, arrowhead in G) of a wild-type embryo under bright-field (G) and dark-field (H). Hnf4 expression in Smad4 mutant embryo under bright-field (I) and dark-field (J) is severely reduced in the visceral endoderm (arrowhead in I). (K) TUNEL staining of wild-type embryo reveals a few apoptotic nuclei in the ectoderm region (large arrowhead) and in the proamniotic cavity (small arrowhead). (L) TUNEL staining of Smad4 mutant embryo indicates a slight increase of apoptosis in the ectoderm (large arrowhead). (M) BrdU staining of wild-type embryo discloses strong BrdU-positive nuclei (arrowhead) throughout the embryo. (N) BrdU staining of Smad4 mutant embryo shows a significantly reduced number of BrdU positive nuclei. (Bar) 60 μm.
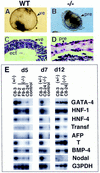
Figure 6
In vitro differentiation of the visceral endoderm and mesoderm is impaired in Smad4 mutant embryoid bodies. Morphological analysis of the EBs at day 10 of culture revealed that most (A) wild-type EBs formed a large cystic cavity. (B) Mutant embryoid bodies gave no sign of cavitation and were smaller in size. (C,D) H+E staining. (C) Wild-type embryoid bodies have a well defined visceral endoderm extending around the cystic cavity. (D) Embryoid bodies from _Smad4_-deficient cells have a discontinuous visceral endoderm and do not form any cystic cavity. (E) Semiquantitative RT–PCR analysis of markers for visceral endoderm and mesoderm development. RNA was extracted from embryoid bodies at day 5, 7, and 12 of in vitro differentiation and analyzed for the expression of early (GATA-4, HNF-1) and late (HNF-4, transferrin, α-fetoprotein) markers for the visceral endoderm development. The expression of Brachyury (T) was examined for mesoderm differentiation. To normalize for the amount of mRNA used as starting material, the cDNA of G3PDH was amplified. Verification of the PCR products was assessed by Southern blot analysis by use of end-labeled primers as probe(s), internal to the sequence amplified. Control RT reactions, without reverse transcriptase, were performed on all RNA preparation for each set of primers used to assess the absence of DNA contamination (data not shown). Only one control RNA from the heterozygous mutant was included for each specific PCR reaction (control lane). Abbreviations. (pre) primitive endoderm, (ect) ectoderm. (Bar) 120 μm in A, 60 μm in B, and 40 μm in C,D).
Figure 7
Wild-type visceral endoderm rescues the gastrulation defect of _Smad4_-deficient embryos. Whole mount in situ hybridization of E8.5 chimeric embryos generated from tetraploid aggregation experiments. (A,B) Embryos derived from Smad4 heterozygous ES cells were hybridized for T, or Krox-20 (K20) and Mox-1. (C,D) Dissected embryos derived from Smad4 homozygous mutant ES clone, C8-24. (C) Less affected embryo with malformation in headfold region (arrowhead). (D) Representative embryo with anterior truncation and a well developed posterior region with somites (arrow). (E,F) T expression (arrow) in embryos derived from Smad4 homozygous mutant ES clone F9-5 and C8-24, respectively. Embryo derived from clone F9-5 is less organized with abnormal head region (arrowhead). (G,H) Embryos derived from Smad4 homozygous mutant ES clone F9-5 and C8-24 respectively, probed with Krox-20 and Mox-1. Arrowhead points to head region. The arrows in B,G, and H point to Mox-1 expression. (Bar) 20 μm.
Similar articles
- Murine gastrulation requires HNF-4 regulated gene expression in the visceral endoderm: tetraploid rescue of Hnf-4(-/-) embryos.
Duncan SA, Nagy A, Chan W. Duncan SA, et al. Development. 1997 Jan;124(2):279-87. doi: 10.1242/dev.124.2.279. Development. 1997. PMID: 9053305 - Differential requirements for Smad4 in TGFbeta-dependent patterning of the early mouse embryo.
Chu GC, Dunn NR, Anderson DC, Oxburgh L, Robertson EJ. Chu GC, et al. Development. 2004 Aug;131(15):3501-12. doi: 10.1242/dev.01248. Epub 2004 Jun 23. Development. 2004. PMID: 15215210 - The tumor suppressor SMAD4/DPC4 is essential for epiblast proliferation and mesoderm induction in mice.
Yang X, Li C, Xu X, Deng C. Yang X, et al. Proc Natl Acad Sci U S A. 1998 Mar 31;95(7):3667-72. doi: 10.1073/pnas.95.7.3667. Proc Natl Acad Sci U S A. 1998. PMID: 9520423 Free PMC article. - Role of Smad4 (DPC4) inactivation in human cancer.
Miyaki M, Kuroki T. Miyaki M, et al. Biochem Biophys Res Commun. 2003 Jul 11;306(4):799-804. doi: 10.1016/s0006-291x(03)01066-0. Biochem Biophys Res Commun. 2003. PMID: 12821112 Review. - DPC4/SMAD4 gene alterations in human cancer, and their functional implications.
Schutte M. Schutte M. Ann Oncol. 1999;10 Suppl 4:56-9. Ann Oncol. 1999. PMID: 10436786 Review.
Cited by
- Fine-tune of intrinsic ERK activity by extrinsic BMP signaling in mouse embryonic stem cells.
Li Z, Chen YG. Li Z, et al. Protein Cell. 2012 Jun;3(6):401-4. doi: 10.1007/s13238-012-2925-5. Epub 2012 Apr 19. Protein Cell. 2012. PMID: 22528752 Free PMC article. - Smad4 is critical for self-renewal of hematopoietic stem cells.
Karlsson G, Blank U, Moody JL, Ehinger M, Singbrant S, Deng CX, Karlsson S. Karlsson G, et al. J Exp Med. 2007 Mar 19;204(3):467-74. doi: 10.1084/jem.20060465. Epub 2007 Mar 12. J Exp Med. 2007. PMID: 17353364 Free PMC article. - Smad4 cooperates with lymphoid enhancer-binding factor 1/T cell-specific factor to increase c-myc expression in the absence of TGF-beta signaling.
Lim SK, Hoffmann FM. Lim SK, et al. Proc Natl Acad Sci U S A. 2006 Dec 5;103(49):18580-5. doi: 10.1073/pnas.0604773103. Epub 2006 Nov 28. Proc Natl Acad Sci U S A. 2006. PMID: 17132729 Free PMC article. - The versatility and paradox of BMP signaling in endothelial cell behaviors and blood vessel function.
Kulikauskas MR, X S, Bautch VL. Kulikauskas MR, et al. Cell Mol Life Sci. 2022 Jan 19;79(2):77. doi: 10.1007/s00018-021-04033-z. Cell Mol Life Sci. 2022. PMID: 35044529 Free PMC article. Review. - Transforming Growth Factor Beta Signaling in Cutaneous Wound Healing: Lessons Learned from Animal Studies.
Finnson KW, Arany PR, Philip A. Finnson KW, et al. Adv Wound Care (New Rochelle). 2013 Jun;2(5):225-237. doi: 10.1089/wound.2012.0419. Adv Wound Care (New Rochelle). 2013. PMID: 24761336 Free PMC article. Review.
References
- Acampora D, Mazan S, Lallemand Y, Avantaggiato V, Maury M, Simeone A, Brulet P. Forebrain and midbrain regions are deleted in Otx2−/− mutants due to a defective anterior neuroectoderm specification during gastrulation. Development. 1995;121:3279–3290. - PubMed
- Ang SL, Rossant J. HNF-3 beta is essential for node and notochord formation in mouse development. Cell. 1994;78:561–574. - PubMed
- Attisano L, Wrana JL. Signal transduction by members of the transforming growth factor-beta superfamily. Cytokine Growth Factor Rev. 1996;7:327–339. - PubMed
- Baker JC, Harland RM. A novel mesoderm inducer, Madr2, functions in the activin signal transduction pathway. Genes & Dev. 1996;10:1880–1889. - PubMed
- Candia AF, Hu J, Crosby J, Lalley PA, Noden D, Nadeau JH, Wright CV. Mox-1 and Mox-2 define a novel homeobox gene subfamily and are differentially expressed during early mesodermal patterning in mouse embryos. Development. 1992;116:1123–1136. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous