The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast grr1p - PubMed (original) (raw)
The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast grr1p
M Ruegger et al. Genes Dev. 1998.
Abstract
Genetic analysis in Arabidopsis has led to the identification of several genes that are required for auxin response. One of these genes, AXR1, encodes a protein related to yeast Aos1p, a protein that functions to activate the ubiquitin-related protein Smt3p. Here we report the identification of a new gene called TRANSPORT INHIBITOR RESPONSE 1 (TIR1). The tir1 mutants are deficient in a variety of auxin-regulated growth processes including hypocotyl elongation and lateral root formation. These results indicate that TIR1 is also required for normal response to auxin. Further, mutations in TIR1 display a synergistic interaction with mutations in AXR1, suggesting that the two genes function in overlapping pathways. The TIR1 protein contains a series of leucine-rich repeats and a recently identified motif called an F box. Sequence comparisons indicate that TIR1 is related to the yeast protein Grr1p and the human protein SKP2. Because Grr1p and other F-box proteins have been implicated in ubiquitin-mediated processes, we speculate that auxin response depends on the modification of a key regulatory protein(s) by ubiquitin or a ubiquitin-related protein.
Figures
Figure 1
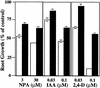
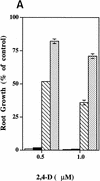
The roots of wild-type (open bars) and tir1-1 (solid bars) seedlings are resistant to the growth-inhibiting properties of NPA, IAA, and 2,4-D. Seeds were germinated on nutrient medium. After 3 days, seedlings were transferred to media containing the indicated compound. Five days later, new root growth was measured and plotted as a percentage of root growth on medium without compound. Bars represent standard errors. Absence of bar indicates error less than thickness of line (n = 12).
Figure 2
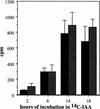
Polar auxin transport is similar in wild-type (solid bars) and tir1-1 (hatched bars) inflorescence stems. Two and one-half centimeters of stem was excised, and the apical end was placed in a nutrient solution containing 1 μ
m
[14C]IAA for the times indicated. The amount of radioactive IAA transported to the basal end of the stem was assayed by liquid scintillation. Each column represents the mean of three replicates; the bar represents the standard error of the mean.
Figure 3
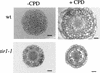
Cell proliferation in response to CPD is reduced in the roots of tir1-1 seedlings. Seedlings were grown for 7 days on nutrient medium plus or minus 5 μ
m
CPD, fixed, and embedded in Spurrs (Ruegger et al. 1997). Root cross sections 300 μm from the root tip are shown. Bars, 20 μm.
Figure 4
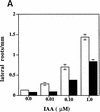
The tir1-1 mutant is deficient in IAA-induction of lateral roots. (A) Eight-millimeter root segments were excised from 5-day-old wild-type (open bar) and tir1-1 (solid bar) seedlings grown on nutrient medium and transferred to medium with IAA. Lateral roots were counted after 5 days by use of a dissecting microscope. Bars represent standard errors. Absence of bar indicates error less than thickness of line (n = 10). (B) Ten-day-old wild-type (left) and tir1-1 (right) seedlings grown on 0.5 μ
m
2,4-D. (Insets) Higher magnification images of roots.
Figure 4
The tir1-1 mutant is deficient in IAA-induction of lateral roots. (A) Eight-millimeter root segments were excised from 5-day-old wild-type (open bar) and tir1-1 (solid bar) seedlings grown on nutrient medium and transferred to medium with IAA. Lateral roots were counted after 5 days by use of a dissecting microscope. Bars represent standard errors. Absence of bar indicates error less than thickness of line (n = 10). (B) Ten-day-old wild-type (left) and tir1-1 (right) seedlings grown on 0.5 μ
m
2,4-D. (Insets) Higher magnification images of roots.
Figure 5
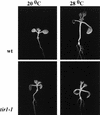
Hypocotyl elongation in response to elevated temperature is reduced in tir1-1 seedlings compared with wild-type seedlings. Seedlings were grown on vertically oriented agar medium for 9 days at the two temperatures.
Figure 6
tir1-1 and axr1-12 have synergistic effects on auxin response and plant morphology. (A) The tir1-1 mutation decreased auxin response in an axr1-12 background at a concentration at which tir1-1 has no effect by itself. Seedlings were grown and treated as described for Fig. 1. (Open bars) Wild type; (solid bars) tir1; (hatched bars) axr1; (stippled bars) axr1 tir1. Bars represent standard error and absence of bar indicates error less than thickness of line. (B) Seedlings (top row) were photographed after 12 days of growth on 1 μ
m
2,4-D. Bar, 2.5 mm. Mature plants (bottom row) were photographed 35 days after germination in soil. Bar, 5 cm.
Figure 6
tir1-1 and axr1-12 have synergistic effects on auxin response and plant morphology. (A) The tir1-1 mutation decreased auxin response in an axr1-12 background at a concentration at which tir1-1 has no effect by itself. Seedlings were grown and treated as described for Fig. 1. (Open bars) Wild type; (solid bars) tir1; (hatched bars) axr1; (stippled bars) axr1 tir1. Bars represent standard error and absence of bar indicates error less than thickness of line. (B) Seedlings (top row) were photographed after 12 days of growth on 1 μ
m
2,4-D. Bar, 2.5 mm. Mature plants (bottom row) were photographed 35 days after germination in soil. Bar, 5 cm.
Figure 7
The characterization of three mutant alleles and transformation rescue of the tir1-1 mutant with a candidate cDNA identify the TIR1 gene. (A) Predicted amino acid sequence of the TIR1 protein. The position of the tir1-1 and tir1-2 mutations, both (G → D), are indicated with an asterisk above the affected residue. The GenBank accession nos. for the cDNA and genomic sequences are AF005048 and AF005047, respectively. (B) RNA blot analysis of TIR1 transcript in tir1 mutants. Twenty-five micrograms of total RNA isolated from 13-day-old seedlings was loaded in each lane. (C) Transformation rescue of the tir1-1 mutant. The p35S:TIR1 plasmid was introduced into tir1-1 plants by vacuum infiltration. Transgenic lines were tested for auxin resistance by plating on medium containing 0.085 μ
m
2,4-D.
Figure 7
The characterization of three mutant alleles and transformation rescue of the tir1-1 mutant with a candidate cDNA identify the TIR1 gene. (A) Predicted amino acid sequence of the TIR1 protein. The position of the tir1-1 and tir1-2 mutations, both (G → D), are indicated with an asterisk above the affected residue. The GenBank accession nos. for the cDNA and genomic sequences are AF005048 and AF005047, respectively. (B) RNA blot analysis of TIR1 transcript in tir1 mutants. Twenty-five micrograms of total RNA isolated from 13-day-old seedlings was loaded in each lane. (C) Transformation rescue of the tir1-1 mutant. The p35S:TIR1 plasmid was introduced into tir1-1 plants by vacuum infiltration. Transgenic lines were tested for auxin resistance by plating on medium containing 0.085 μ
m
2,4-D.
Figure 8
The TIR1 protein has an F box and 16 LRRs. (A) Alignment of F-box motifs from diverse proteins. Identical residues are boxed. (B) Alignment of LRRs in TIR1. The affected residues in the tir1-1 and tir1-2 mutants are underlined. In consensus sequences listed in A and B, aliphatic residues are indicated with an a. (C) Comparison of TIR1, SKP2, Grr1, and C02F5.7. Boxes represent the LRRs.
Similar articles
- Mutations in the TIR1 auxin receptor that increase affinity for auxin/indole-3-acetic acid proteins result in auxin hypersensitivity.
Yu H, Moss BL, Jang SS, Prigge M, Klavins E, Nemhauser JL, Estelle M. Yu H, et al. Plant Physiol. 2013 May;162(1):295-303. doi: 10.1104/pp.113.215582. Epub 2013 Mar 28. Plant Physiol. 2013. PMID: 23539280 Free PMC article. - Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins.
Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. Gray WM, et al. Nature. 2001 Nov 15;414(6861):271-6. doi: 10.1038/35104500. Nature. 2001. PMID: 11713520 - Arabidopsis SGT1b is required for SCF(TIR1)-mediated auxin response.
Gray WM, Muskett PR, Chuang HW, Parker JE. Gray WM, et al. Plant Cell. 2003 Jun;15(6):1310-9. doi: 10.1105/tpc.010884. Plant Cell. 2003. PMID: 12782725 Free PMC article. - Auxin receptors: a new role for F-box proteins.
Parry G, Estelle M. Parry G, et al. Curr Opin Cell Biol. 2006 Apr;18(2):152-6. doi: 10.1016/j.ceb.2006.02.001. Epub 2006 Feb 17. Curr Opin Cell Biol. 2006. PMID: 16488128 Review. - Function of the ubiquitin-proteasome pathway in auxin response.
Gray WM, Estelle I. Gray WM, et al. Trends Biochem Sci. 2000 Mar;25(3):133-8. doi: 10.1016/s0968-0004(00)01544-9. Trends Biochem Sci. 2000. PMID: 10694884 Review.
Cited by
- Ethylene- and Shade-Induced Hypocotyl Elongation Share Transcriptome Patterns and Functional Regulators.
Das D, St Onge KR, Voesenek LA, Pierik R, Sasidharan R. Das D, et al. Plant Physiol. 2016 Oct;172(2):718-733. doi: 10.1104/pp.16.00725. Epub 2016 Jun 21. Plant Physiol. 2016. PMID: 27329224 Free PMC article. - Conditional Degradation of UNC-31/CAPS Enables Spatiotemporal Analysis of Neuropeptide Function.
Cornell R, Cao W, Liu J, Pocock R. Cornell R, et al. J Neurosci. 2022 Nov 16;42(46):8599-8607. doi: 10.1523/JNEUROSCI.1368-22.2022. Epub 2022 Oct 27. J Neurosci. 2022. PMID: 36302635 Free PMC article. - Regulated proteolysis and plant development.
Schwechheimer C, Schwager K. Schwechheimer C, et al. Plant Cell Rep. 2004 Nov;23(6):353-64. doi: 10.1007/s00299-004-0858-z. Epub 2004 Sep 10. Plant Cell Rep. 2004. PMID: 15365760 Review. - An auxin-inducible F-box protein CEGENDUO negatively regulates auxin-mediated lateral root formation in Arabidopsis.
Dong L, Wang L, Zhang Y, Zhang Y, Deng X, Xue Y. Dong L, et al. Plant Mol Biol. 2006 Mar;60(4):599-615. doi: 10.1007/s11103-005-5257-5. Plant Mol Biol. 2006. PMID: 16525894 - Odyssey of auxin.
Abel S, Theologis A. Abel S, et al. Cold Spring Harb Perspect Biol. 2010 Oct;2(10):a004572. doi: 10.1101/cshperspect.a004572. Epub 2010 Jan 27. Cold Spring Harb Perspect Biol. 2010. PMID: 20739413 Free PMC article. Review.
References
- Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. - PubMed
- Barbier-Brygoo H. Tracking auxin receptors using functional approaches. Crit Rev Plant Sci. 1995;14:1–25.
- Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. CR Acad Sci Ser III Sci Vie. 1993;316:1194–1199.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous