p300 and ATF-2 are components of the DRF complex, which regulates retinoic acid- and E1A-mediated transcription of the c-jun gene in F9 cells - PubMed (original) (raw)
p300 and ATF-2 are components of the DRF complex, which regulates retinoic acid- and E1A-mediated transcription of the c-jun gene in F9 cells
H Kawasaki et al. Genes Dev. 1998.
Abstract
Transcriptional activation of the c-jun gene is a critical event in the differentiation of F9 cells. In our previous studies we characterized an element [differentiation response element (DRE)] in the c-jun promoter that is both necessary and sufficient to confer the capacity for differentiation-dependent up-regulation. This element binds the differentiation regulatory factor (DRF) complex, of which one component is the adenovirus E1A-associated protein p300. We have now identified activation transcription factor-2 (ATF-2) as a DNA-binding subunit of the DRF complex. p300 and ATF-2 interact with each other in vivo and in vitro. The bromodomain and the C/H2 domain of p300 mediate the binding to ATF-2, which in turn requires a proline-rich region between amino acids 112 and 350 for its interaction with p300. The phosphorylation of the serine residue at position 121 of ATF-2 appears to be induced by protein kinase C alpha (PKC alpha) after treatment of cells with retinoic acid (RA) or induction with E1A. In cotransfection assays, wild-type ATF-2 enhanced the transcription of an E2/tk-luciferase construct, in conjunction with p300-E2. However, a mutant form of ATF-2 with a mutation at position 121 (pCMVATF-2(Ser121-Ala)) did not. These results suggest that ATF-2 and p300 cooperate in the control of transcription by forming a protein complex that is responsive to differentiation-inducing signals, such as RA or E1A, and moreover, that the phosphorylation of ATF-2 by PKC alpha is probably a signaling event in the pathway that leads to the transactivation of the c-jun gene in F9 cells.
Figures
Figure 1
Selective interaction between p300 and ATF-2 protein species in vivo and the effects of phosphatase in the electrophoretic mobilities of ATF-2. (A) Cell lysates from F9 cells and RA-treated F9 cells were immunoprecipitated directly with antibodies specific for p300 (α-p300; RW128) or were subjected to sequential immunoprecipitations with antibodies against ATF-2 (α-ATF-2; C-19) and antibodies against p300, as described in Materials and Methods. The immunoprecipitated proteins were resolved by SDS-PAGE (5% polyacrylamide). (Lanes 1,3,5,7) Extracts of F9 cells; (lanes 2,4,6,8) p300 from extracts of differentiated F9 cells. (Lanes 1–4) [35S]Methionine-labeled p300; (lanes 5–8) 32P-labeled p300. The positions of both bands of p300 proteins are indicated by arrows. (B) Cell lysates from F9 cells (lane 1) and RA-treated F9 cells (lanes 2–4) were incubated with 50 units of CIAP at 37°C for 30 min in 50 m
m
Tris-HCl (pH 8.0) in the presence (lane 4) or absence (lane 3) of phosphatase inhibitors (10 m
m
sodium phosphate, 15 m
m
sodium pyrophosphate, 5 m
m
NaF, 0.1 m
m
Na3VO4). The immunoprecipitated proteins were resolved by SDS-PAGE (10% polyacrylamide) and immunoblotted with antibodies specific for ATF-2 (C-19). The positions of bands of ATF-2 are indicated by arrows.
Figure 1
Selective interaction between p300 and ATF-2 protein species in vivo and the effects of phosphatase in the electrophoretic mobilities of ATF-2. (A) Cell lysates from F9 cells and RA-treated F9 cells were immunoprecipitated directly with antibodies specific for p300 (α-p300; RW128) or were subjected to sequential immunoprecipitations with antibodies against ATF-2 (α-ATF-2; C-19) and antibodies against p300, as described in Materials and Methods. The immunoprecipitated proteins were resolved by SDS-PAGE (5% polyacrylamide). (Lanes 1,3,5,7) Extracts of F9 cells; (lanes 2,4,6,8) p300 from extracts of differentiated F9 cells. (Lanes 1–4) [35S]Methionine-labeled p300; (lanes 5–8) 32P-labeled p300. The positions of both bands of p300 proteins are indicated by arrows. (B) Cell lysates from F9 cells (lane 1) and RA-treated F9 cells (lanes 2–4) were incubated with 50 units of CIAP at 37°C for 30 min in 50 m
m
Tris-HCl (pH 8.0) in the presence (lane 4) or absence (lane 3) of phosphatase inhibitors (10 m
m
sodium phosphate, 15 m
m
sodium pyrophosphate, 5 m
m
NaF, 0.1 m
m
Na3VO4). The immunoprecipitated proteins were resolved by SDS-PAGE (10% polyacrylamide) and immunoblotted with antibodies specific for ATF-2 (C-19). The positions of bands of ATF-2 are indicated by arrows.
Figure 2
ATF-2 binds the DRE that responds to p300 in differentiated and undifferentiated F9 cells. (A) Nuclear extracts from F9 cells were incubated on ice for 30 min without (lanes 1,2) or with (lanes 3–14) various antibodies and then analyzed in a band-shift assay with DRE as the probe. (Lane 1) Free DRE probe (F); (lane 2) nuclear extract alone; (lane 3) antibodies against ATF-2 (α-ATF-2; Maekawa et al. 1989); (lane 4) antibodies against ATF-2 that had been preabsorbed with 3 μg of GST–ATF-2; (lane 6) antibodies against ATF-2 (Livingston et al. 1995); (lanes 5,7–14) various antibodies were added to the reactions as indicated (α-n, antibodies against protein n). The positions of bands of DRF1 and DRF2 are indicated by arrows. (B) Nuclear extracts of RA-treated F9 cells were incubated on ice without (lanes 1,2) or with (lane 5) 4 μg of rabbit IgG or with various antibodies (lanes 3,4,6–14) for 30 min and analyzed in a band-shift assay with DRE as the probe. (Lane 1) Free DRE probe (F); (lane 2) nuclear extract alone; (lane 3) antibodies against ATF-2 (Maekawa et al. 1989); (lane 4) antibodies against ATF-2 that had been preabsorbed with 3 μg of GST–ATF-2; (lanes 6–14) various antibodies were added to the reactions as indicated. The positions of bands of DRF1 are indicated by arrows. (C) Nuclear extracts from RA-treated F9 cells were incubated on ice without (lane 1) or with 10−2 dilution (lane 2) or 10−1 dilution (lane 3) of antiserum against ATF-2 (Maekawa et al. 1989; Livingston et al. 1995); with 10−1 dilution of antiserum against c-Jun (lane 4); or with 25-fold molar excess of DRE oligonucleotide as competitor (lane 5; Kitabayashi et al. 1995) for 30 min; then proteins were analyzed in the band-shift assay with DRE as the probe. The positions of bands of DRF1 are indicated by arrows. (F) Free DRE DNA probe.
Figure 2
ATF-2 binds the DRE that responds to p300 in differentiated and undifferentiated F9 cells. (A) Nuclear extracts from F9 cells were incubated on ice for 30 min without (lanes 1,2) or with (lanes 3–14) various antibodies and then analyzed in a band-shift assay with DRE as the probe. (Lane 1) Free DRE probe (F); (lane 2) nuclear extract alone; (lane 3) antibodies against ATF-2 (α-ATF-2; Maekawa et al. 1989); (lane 4) antibodies against ATF-2 that had been preabsorbed with 3 μg of GST–ATF-2; (lane 6) antibodies against ATF-2 (Livingston et al. 1995); (lanes 5,7–14) various antibodies were added to the reactions as indicated (α-n, antibodies against protein n). The positions of bands of DRF1 and DRF2 are indicated by arrows. (B) Nuclear extracts of RA-treated F9 cells were incubated on ice without (lanes 1,2) or with (lane 5) 4 μg of rabbit IgG or with various antibodies (lanes 3,4,6–14) for 30 min and analyzed in a band-shift assay with DRE as the probe. (Lane 1) Free DRE probe (F); (lane 2) nuclear extract alone; (lane 3) antibodies against ATF-2 (Maekawa et al. 1989); (lane 4) antibodies against ATF-2 that had been preabsorbed with 3 μg of GST–ATF-2; (lanes 6–14) various antibodies were added to the reactions as indicated. The positions of bands of DRF1 are indicated by arrows. (C) Nuclear extracts from RA-treated F9 cells were incubated on ice without (lane 1) or with 10−2 dilution (lane 2) or 10−1 dilution (lane 3) of antiserum against ATF-2 (Maekawa et al. 1989; Livingston et al. 1995); with 10−1 dilution of antiserum against c-Jun (lane 4); or with 25-fold molar excess of DRE oligonucleotide as competitor (lane 5; Kitabayashi et al. 1995) for 30 min; then proteins were analyzed in the band-shift assay with DRE as the probe. The positions of bands of DRF1 are indicated by arrows. (F) Free DRE DNA probe.
Figure 3
Interaction of in vitro-translated ATF-2 with deletion variants of GST–p300. (A) Schematic representation of the variants of GST–p300. Shown are p300 and deletion derivatives fused to the GST protein. The patterned boxes represent the cysteine/histidine-rich regions C/H1, C/H2, and C/H3, and are labeled; (Br) The bromodomain. The numbers at right indicate the amino acids of p300. (FL p300) Full-length p300 protein; [p300Δ(amino acids 963–1571)], p300 protein lacking amino acids 963–1571; (GST) glutathione _S_-transferase–truncated protein. (B) In vitro translated [35S]methionine-labeled ATF-2 was incubated with the GST–p300 variants that consisted of the amino-terminal, carboxy-terminal, and central portions of the protein (lanes 5–19) or variants that lacked amino acids 963–1571 (lanes 20–22), or ATF-2 was incubated with GST alone (lanes 2–4). The bound ATF-2 is indicated by an arrow at left. (Lane 1) Input, namely, in vitro-translated [35S]methionine-labeled ATF-2. The input lane contained 1.5% (in terms of cpm) of the radiolabeled protein used in the binding experiments.
Figure 3
Interaction of in vitro-translated ATF-2 with deletion variants of GST–p300. (A) Schematic representation of the variants of GST–p300. Shown are p300 and deletion derivatives fused to the GST protein. The patterned boxes represent the cysteine/histidine-rich regions C/H1, C/H2, and C/H3, and are labeled; (Br) The bromodomain. The numbers at right indicate the amino acids of p300. (FL p300) Full-length p300 protein; [p300Δ(amino acids 963–1571)], p300 protein lacking amino acids 963–1571; (GST) glutathione _S_-transferase–truncated protein. (B) In vitro translated [35S]methionine-labeled ATF-2 was incubated with the GST–p300 variants that consisted of the amino-terminal, carboxy-terminal, and central portions of the protein (lanes 5–19) or variants that lacked amino acids 963–1571 (lanes 20–22), or ATF-2 was incubated with GST alone (lanes 2–4). The bound ATF-2 is indicated by an arrow at left. (Lane 1) Input, namely, in vitro-translated [35S]methionine-labeled ATF-2. The input lane contained 1.5% (in terms of cpm) of the radiolabeled protein used in the binding experiments.
Figure 4
Interaction of GST–p300 with deletion variants of ATF-2. (A) Schematic representation of the ATF-2 fusion proteins synthesized in insect cells. (B) Extracts of infected insect cells containing wild-type or mutant ATF-2 proteins were incubated with immobilized GST–p300 fusion protein for 2 hr at 4°C. The beads were washed extensively and bound protein was analyzed by Western blotting analysis with antibodies specific for the amino- or carboxy-terminal-specific region of ATF-2 (lanes 3,6,9,12,15). Beads with bound GST protein (lanes 2,5,8,11,14) were used as a control. Lanes 1,4,7,10, and 13 (Load) were loaded with 0.5% (in terms of cpm) of the input recombinant ATF-2 or the variant proteins used in the binding experiments. The positions of ATF-2–FL, ATF-2–ΔN, ATF-2–Δ21, ATF-2–Δ9 and ATF-2-Δ(amino acids 112–350) are indicated by arrows.
Figure 4
Interaction of GST–p300 with deletion variants of ATF-2. (A) Schematic representation of the ATF-2 fusion proteins synthesized in insect cells. (B) Extracts of infected insect cells containing wild-type or mutant ATF-2 proteins were incubated with immobilized GST–p300 fusion protein for 2 hr at 4°C. The beads were washed extensively and bound protein was analyzed by Western blotting analysis with antibodies specific for the amino- or carboxy-terminal-specific region of ATF-2 (lanes 3,6,9,12,15). Beads with bound GST protein (lanes 2,5,8,11,14) were used as a control. Lanes 1,4,7,10, and 13 (Load) were loaded with 0.5% (in terms of cpm) of the input recombinant ATF-2 or the variant proteins used in the binding experiments. The positions of ATF-2–FL, ATF-2–ΔN, ATF-2–Δ21, ATF-2–Δ9 and ATF-2-Δ(amino acids 112–350) are indicated by arrows.
Figure 5
ATF-2 and p300 cooperate to transactivate the c-jun promoter in F9 cells. F9 cells that had been stably transfected with 5 μg of −730/+874 c-Jun CAT (wild type; lanes 1–17,25–30,33,34) and 5 μg of −730/+874 c-Jun CAT–m182/175 (mDRE; lanes 18–24,31,32) were cotransfected with 4 μg of pCMV–ATF-1 (lanes 6–9), pCMV–ATF-2 (lanes 10–13), pCMV–ATF-2 Δ(amino acids 112–350) (lanes 25–27) or pCMV–ATF-3 (lanes 14–17) in the presence of the pACT–p300 expression plasmid (lanes 6–17,21–27) or pACT–p300 Δ(amino acids 963–1571) (lanes 28–30), or pCMV-E1A (lanes 32,34) plus 2 μg of pRSVLacZ (lanes 1–34), as indicated. The cells were incubated for 72 hr and CAT assays were performed with extracts in which the amount of protein had been normalized by the activity of β-galactosidase. In some cases, F9 cells were incubated with 3 × 10−7
m
RA (lanes 31,33). The percent conversion of CAT to its acetylated form is indicated. (Lane 1) Wild type only; (lanes 2–4,18–20) 4 μg of pCMV–ATF-1, pCMV–ATF-2, or pCMV–ATF-3; (lane 5) 4 μg of pACT–p300; (lanes 6–9,10–13,14–17,25–30) 1.0, 4.0, and 8.0 μg of pCMV–ATF-1, pCMV–ATF-2, pCMV–ATF-2 Δ(amino acids 112–350) or pCMV–ATF-3 plasmid, respectively, in the presence of 4 μg of pACT–p300 (lanes 6–17,25–27) or of pACT–p300 Δ(amino acids 965–1571) (lanes 28–30), respectively.
Figure 6
ATF-2 and p300 cooperate to transactivate DRE-mediated transcription in F9 cells. F9 cells that had been stably transfected with 5 μg of 3× DRE/tk–CAT (lanes 1–4,9–11,15–22) or tk–CAT (lanes 5–8,12–14) were cotransfected with various amounts of pCMV–ATF-2 (lanes 2–4,6–8), pCMV–ATF-2 Δ(amino acids 112–350) (lanes 18–20), pACT–p300 (lanes 9–11,12–14), or pACT–p300 Δ(amino acids 963–1571) (lanes 15–17) and 5 μg (lanes 9–11,15–17), 8 μg (lanes 12–14) of pCMV–ATF-2, or 2 μg of pRSV–LacZ (lanes 1–22). The cells were incubated for 72 hr and CAT assays were performed with extracts in which the amount of protein had been normalized by the activity of β-galactosidase. The percent conversion of CAT to its acetylated forms is indicated. (Lane 1) DRE/tk–CAT only; (lanes 2–4,6–8,18–20) 1.0, 4.0, and 8.0 μg of pCMV–ATF-2 or pCMV–ATF-2 Δ(amino acids 112–350), respectively; (lane 5) tk–CAT only; (lanes 9–17) 1.0, 4.0, and 8.0 μg of pACT–p300 or pACT–p300 Δ(amino acids 963–1571), respectively. (Lane 21) treatment with 3 × 10−7
m
RA; (lane 22) transfection with pCMV–E1A (4 μg).
Figure 7
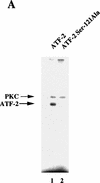
ATF-2 is phosphorylated by PKC and activates a p300–E2 fusion protein. (A) Five micrograms of ATF-2 and mutant ATF-2Ser121–Ala proteins were purified from E. coli and incubated with 25 ng of a mixture of recombinant PKCα, PKCβ, and PKCγ mixture in the appropriate kinase buffer (see Materials and Methods) for 10 min at 30°C. The resultant phosphorylated proteins were analyzed by SDS-PAGE (10% polyacrylamide). (Lane 1) ATF-2; (lane 2) ATF-2Ser121–Ala mutant protein. (B,C) F9 cells were incubated with 3 × 10−7
m
RA for the indicated periods of time. Nuclei were isolated and nuclear lysates were subjected to immunoprecipitation with polyclonal antibodies specific for PKCα (B) and PKCβ (C), respectively, and protein A–Sepharose for 2 hr at 4°C. The immune complexes were incubated with 5 μCi of [γ-32P]ATP in kinase buffer for 30 min at 30°C. The resultant phosphorylated protein complexes were subjected to SDS-PAGE (10% polyacrylamide). (Lane 1) F9 cell extract; (lanes 2–4) F9 cells lysate that had been incubated with RA for 9, 18, and 72 hr, respectively. The immunoprecipitates with antibodies specific for PKCα or PKCβ were immunoblotted with the antibodies against the respective PKC (bottom panels). (D) F9 cells that had been stably transfected with 5 μg of tk–luc or 5× E2/tk–luc were cotransfected with 3 μg of pCMV–p300–E2 with or without 4 μg of pCMV–ATF-2 or 4 μg of pCMV–ATF-2Ser121–Ala plus 2 μg of pRSV-LacZ, as indicated. The cells were incubated for 72 hr in the absence or presence of 3 × 10−7
m
RA or 2.5 μg of pCMV–E1A. The results are expressed as the extent of induction (_x_-fold) as compared to the results with the control plasmid (pcDNA3). The results are the means from three independent experiments.
Figure 7
ATF-2 is phosphorylated by PKC and activates a p300–E2 fusion protein. (A) Five micrograms of ATF-2 and mutant ATF-2Ser121–Ala proteins were purified from E. coli and incubated with 25 ng of a mixture of recombinant PKCα, PKCβ, and PKCγ mixture in the appropriate kinase buffer (see Materials and Methods) for 10 min at 30°C. The resultant phosphorylated proteins were analyzed by SDS-PAGE (10% polyacrylamide). (Lane 1) ATF-2; (lane 2) ATF-2Ser121–Ala mutant protein. (B,C) F9 cells were incubated with 3 × 10−7
m
RA for the indicated periods of time. Nuclei were isolated and nuclear lysates were subjected to immunoprecipitation with polyclonal antibodies specific for PKCα (B) and PKCβ (C), respectively, and protein A–Sepharose for 2 hr at 4°C. The immune complexes were incubated with 5 μCi of [γ-32P]ATP in kinase buffer for 30 min at 30°C. The resultant phosphorylated protein complexes were subjected to SDS-PAGE (10% polyacrylamide). (Lane 1) F9 cell extract; (lanes 2–4) F9 cells lysate that had been incubated with RA for 9, 18, and 72 hr, respectively. The immunoprecipitates with antibodies specific for PKCα or PKCβ were immunoblotted with the antibodies against the respective PKC (bottom panels). (D) F9 cells that had been stably transfected with 5 μg of tk–luc or 5× E2/tk–luc were cotransfected with 3 μg of pCMV–p300–E2 with or without 4 μg of pCMV–ATF-2 or 4 μg of pCMV–ATF-2Ser121–Ala plus 2 μg of pRSV-LacZ, as indicated. The cells were incubated for 72 hr in the absence or presence of 3 × 10−7
m
RA or 2.5 μg of pCMV–E1A. The results are expressed as the extent of induction (_x_-fold) as compared to the results with the control plasmid (pcDNA3). The results are the means from three independent experiments.
Figure 7
ATF-2 is phosphorylated by PKC and activates a p300–E2 fusion protein. (A) Five micrograms of ATF-2 and mutant ATF-2Ser121–Ala proteins were purified from E. coli and incubated with 25 ng of a mixture of recombinant PKCα, PKCβ, and PKCγ mixture in the appropriate kinase buffer (see Materials and Methods) for 10 min at 30°C. The resultant phosphorylated proteins were analyzed by SDS-PAGE (10% polyacrylamide). (Lane 1) ATF-2; (lane 2) ATF-2Ser121–Ala mutant protein. (B,C) F9 cells were incubated with 3 × 10−7
m
RA for the indicated periods of time. Nuclei were isolated and nuclear lysates were subjected to immunoprecipitation with polyclonal antibodies specific for PKCα (B) and PKCβ (C), respectively, and protein A–Sepharose for 2 hr at 4°C. The immune complexes were incubated with 5 μCi of [γ-32P]ATP in kinase buffer for 30 min at 30°C. The resultant phosphorylated protein complexes were subjected to SDS-PAGE (10% polyacrylamide). (Lane 1) F9 cell extract; (lanes 2–4) F9 cells lysate that had been incubated with RA for 9, 18, and 72 hr, respectively. The immunoprecipitates with antibodies specific for PKCα or PKCβ were immunoblotted with the antibodies against the respective PKC (bottom panels). (D) F9 cells that had been stably transfected with 5 μg of tk–luc or 5× E2/tk–luc were cotransfected with 3 μg of pCMV–p300–E2 with or without 4 μg of pCMV–ATF-2 or 4 μg of pCMV–ATF-2Ser121–Ala plus 2 μg of pRSV-LacZ, as indicated. The cells were incubated for 72 hr in the absence or presence of 3 × 10−7
m
RA or 2.5 μg of pCMV–E1A. The results are expressed as the extent of induction (_x_-fold) as compared to the results with the control plasmid (pcDNA3). The results are the means from three independent experiments.
Similar articles
- The coactivators p300 and CBP have different functions during the differentiation of F9 cells.
Ugai H, Uchida K, Kawasaki H, Yokoyama KK. Ugai H, et al. J Mol Med (Berl). 1999 Jun;77(6):481-94. doi: 10.1007/s001099900021. J Mol Med (Berl). 1999. PMID: 10475063 Review. - Phosphorylation of the adenovirus E1A-associated 300 kDa protein in response to retinoic acid and E1A during the differentiation of F9 cells.
Kitabayashi I, Eckner R, Arany Z, Chiu R, Gachelin G, Livingston DM, Yokoyama KK. Kitabayashi I, et al. EMBO J. 1995 Jul 17;14(14):3496-509. doi: 10.1002/j.1460-2075.1995.tb07356.x. EMBO J. 1995. PMID: 7628451 Free PMC article. - JDP2, a repressor of AP-1, recruits a histone deacetylase 3 complex to inhibit the retinoic acid-induced differentiation of F9 cells.
Jin C, Li H, Murata T, Sun K, Horikoshi M, Chiu R, Yokoyama KK. Jin C, et al. Mol Cell Biol. 2002 Jul;22(13):4815-26. doi: 10.1128/MCB.22.13.4815-4826.2002. Mol Cell Biol. 2002. PMID: 12052888 Free PMC article. - The N-terminal transactivation domain of ATF2 is a target for the co-operative activation of the c-jun promoter by p300 and 12S E1A.
Duyndam MC, van Dam H, Smits PH, Verlaan M, van der Eb AJ, Zantema A. Duyndam MC, et al. Oncogene. 1999 Apr 8;18(14):2311-21. doi: 10.1038/sj.onc.1202584. Oncogene. 1999. PMID: 10327051 - Synergistic role of E1A-binding proteins and tissue-specific transcription factors in differentiation.
Condorelli G, Giordano A. Condorelli G, et al. J Cell Biochem. 1997 Dec 15;67(4):423-31. doi: 10.1002/(sici)1097-4644(19971215)67:4<423::aid-jcb1>3.0.co;2-u. J Cell Biochem. 1997. PMID: 9383702 Review.
Cited by
- The N-terminal domain of p73 interacts with the CH1 domain of p300/CREB binding protein and mediates transcriptional activation and apoptosis.
Zeng X, Li X, Miller A, Yuan Z, Yuan W, Kwok RP, Goodman R, Lu H. Zeng X, et al. Mol Cell Biol. 2000 Feb;20(4):1299-310. doi: 10.1128/MCB.20.4.1299-1310.2000. Mol Cell Biol. 2000. PMID: 10648616 Free PMC article. - Suppressor role of activating transcription factor 2 (ATF2) in skin cancer.
Bhoumik A, Fichtman B, Derossi C, Breitwieser W, Kluger HM, Davis S, Subtil A, Meltzer P, Krajewski S, Jones N, Ronai Z. Bhoumik A, et al. Proc Natl Acad Sci U S A. 2008 Feb 5;105(5):1674-9. doi: 10.1073/pnas.0706057105. Epub 2008 Jan 28. Proc Natl Acad Sci U S A. 2008. PMID: 18227516 Free PMC article. - Upregulation of COX-2/PGE2 by ET-1 mediated through Ca2+-dependent signals in mouse brain microvascular endothelial cells.
Lin CC, Hsieh HL, Chi PL, Yang CC, Hsiao LD, Yang CM. Lin CC, et al. Mol Neurobiol. 2014 Jun;49(3):1256-69. doi: 10.1007/s12035-013-8597-1. Epub 2013 Nov 28. Mol Neurobiol. 2014. PMID: 24287977 - Signaling pathways mediating beta3-adrenergic receptor-induced production of interleukin-6 in adipocytes.
Tchivileva IE, Tan KS, Gambarian M, Nackley AG, Medvedev AV, Romanov S, Flood PM, Maixner W, Makarov SS, Diatchenko L. Tchivileva IE, et al. Mol Immunol. 2009 Jul;46(11-12):2256-66. doi: 10.1016/j.molimm.2009.04.008. Epub 2009 May 23. Mol Immunol. 2009. PMID: 19477016 Free PMC article. - A CBP/p300 homolog specifies multiple differentiation pathways in Caenorhabditis elegans.
Shi Y, Mello C. Shi Y, et al. Genes Dev. 1998 Apr 1;12(7):943-55. doi: 10.1101/gad.12.7.943. Genes Dev. 1998. PMID: 9531533 Free PMC article.
References
- Abraham SE, Lobo S, Yaciuk P, Wang H-GH, Moran E. p300, and p300-associated proteins, are components of TATA-binding protein (TBP) complexes. Oncogene. 1993;8:1639–1647. - PubMed
- Arany Z, Seller WR, Livingston DM, Eckner R. E1A-associated p300 and CREB-associated CBP belong to a conserved family of coactivators. Cell. 1994;77:799–800. - PubMed
- Arany Z, Newsome D, Oldread E, Livingston DM, Eckner R. A family of transcriptional adapter proteins targeted by the E1A oncoprotein. Nature. 1995;374:81–84. - PubMed
- Avantaggiati ML, Ogryzko V, Gardner K, Giordano A, Levine AS, Kelly K. Recruitment of p300/CBP in p53-dependent signal pathways. Cell. 1997;89:1175–1184. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous