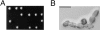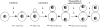Sequential assembly of myosin II, an IQGAP-like protein, and filamentous actin to a ring structure involved in budding yeast cytokinesis - PubMed (original) (raw)
Sequential assembly of myosin II, an IQGAP-like protein, and filamentous actin to a ring structure involved in budding yeast cytokinesis
J Lippincott et al. J Cell Biol. 1998.
Abstract
We have identified a Saccharomyces cerevisiae protein, Cyk1p, that exhibits sequence similarity to the mammalian IQGAPs. Gene disruption of Cyk1p results in a failure in cytokinesis without affecting other events in the cell cycle. Cyk1p is diffused throughout most of the cell cycle but localizes to a ring structure at the mother-bud junction after the initiation of anaphase. This ring contains filamentous actin and Myo1p, a myosin II homologue. In vivo observation with green fluorescent protein-tagged Myo1p showed that the ring decreases drastically in size during cell division and therefore may be contractile. These results indicate that cytokinesis in budding yeast is likely to involve an actomyosin-based contractile ring. The assembly of this ring occurs in temporally distinct steps: Myo1p localizes to a ring that overlaps the septins at the G1-S transition slightly before bud emergence; Cyk1p and actin then accumulate in this ring after the activation of the Cdc15 pathway late in mitosis. The localization of myosin is abolished by a mutation in Cdc12p, implicating a role for the septin filaments in the assembly of the actomyosin ring. The accumulation of actin in the cytokinetic ring was not observed in cells depleted of Cyk1p, suggesting that Cyk1p plays a role in the recruitment of actin filaments, perhaps through a filament-binding activity similar to that demonstrated for mammalian IQGAPs.
Figures
Figure 3
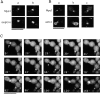
Cyk1p is required for cytokinesis. (A) RLY261 (a and c; wild-type) and RLY277 (b and d; expressing Cyk1 under GAL1 promoter) were streaked onto a YPG plate (a and b) and a YPD plate (c and d), and then grown for 3 d at 30°C before photography. (B) RLY261 and RLY277 cells were cultured overnight in YPG at 30°C. A 10-ml sample of RLY277 culture was processed for immunoblot analysis (lane 1). The rest of the cells were shifted to YPD containing 0.1 μg/ml α factor (Sigma Chemical Co., St. Louis, MO) and grown for 3 h at 30°C. The cells were washed five times with water and transferred to YPD. 10-ml samples were taken at 0, 45, 60, 75, and 90 min (lane 2, 3, 4, 5, and 6, respectively) after the release. Immunoblot analysis using anti-myc antibody was carried out as described in Materials and Methods. (C) In the same experiment as in B, 5-ml samples were fixed with formaldehyde at each time point after the release from the G1 arrest (horizontal axis in each graph). A fraction of the cells were treated with zymolyase and stained with DAPI and rhodamine phalloidin. The number of budded cells or cells with divided DNA mass was counted and divided by total number of cells counted to give percent budded cells (% budded) (a), and percent with divided DNA (% with divided DNA) (b), respectively. The number of cells in which actin patches were concentrated at the mother–bud junction or with an actin bar at the mother–bud junction was divided by the total number of cells counted (typically 100–150) to give percent with patches at septum or percent with an actin bar at septum (d), respectively. Another fraction of the fixed cells from each time point was sonicated and counted on a hemocytometer. The resultant cell concentration at each time point was divided by that at time 0 to give relative cell number (c). In a, b, and c: ○, wild type; ▪, Cyk1− cells. ▴ in a, Cyk1− cells with more than one bud. Asterisk in c, relative cell number after zymolyase treatment. ○ in d, wild-type cells with actin patches at septum; ▵, wild-type cells with actin bar at septum; •, Cyk1− cells with actin patches at septum; ▴, Cyk1− cells with actin bar at septum. (D) Cyk1− cells after 4 h in glucose from the same experiment as in B and C were fixed, treated with zymolyase, and then stained with DAPI. A representative field of cells is shown. (E) Cyk1− cells from the 135-min time point were processed for thin section electron microscopy. Examples of cells with two buds connected to the mother cell are shown. (F) Cyk1− cells from the 75-min time point were stained with rhodamine phalloidin. A representative field of cells is shown. Bars: (D and F) 10 μm.; (E) 1 μm.
Figure 1
Comparison of structural domains of Cyk1p with human IQGAP1. The complete amino acid sequence of Cyk1 protein is not shown but these data are available from GenBank/ EMBL/DDBJ under accession number Z73598; and yeast genome database ORF name: YPL242c). (A) Schematic diagram comparing the domain organization of human IQGAP1 (these data are available from GenBank/EMBL/DDBJ under accession number L33075) and Cyk1p. CHD, calponin homology domain; WW, WW domain; IQ, IQ motifs; and GRD, GAP-related domain. (B) Alignment of the calponin homology domains of Cyk1p and IQGAP1 with the corresponding segment of mouse calponin (Swiss Protein database accession number: Q08093). Identical residues are shaded in black, conserved substitutions are shaded in gray. (C) Alignment of the GRDs of IQGAP1 and Cyk1p with the catalytic domain of Schizosaccharomyces pombe Ras GAP Sar1 (available from GenBank/EMBL/DDBJ accession number S37449). Identical residues are shaded in black, conserved substitutions are shaded in gray. (D) Alignment of the eight IQ motifs in Cyk1p. The consensus residues are shaded in black.
Figure 2
Phenotype of CYK1 gene disruption. (A) Tetrad analysis of a diploid strain (RLY226) heterozygous for the Δcyk1 mutation showing 2:2 segregation of viability. The plate was photographed after 5-d growth at 23°C. (B) The morphology of a typical Δcyk1 microcolony after 24-h growth at 23°C, after tetrad dissection. The microcolony did not further increase in size and the cells eventually lysed. The photograph was taken under a Zeiss Axiophot microscope with a ×40 Nikon 0.5 ELWD objective. Bar, 20 μm.
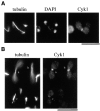
Figure 4
Cyk1p colocalizes with an actin ring. RLY230 (Cyk1-myc–expressing) cells were grown in YPD and fixed with formaldehyde. (A) A representative field of cells stained with mouse anti-myc antibody. (B) Examples of cells that (a) show a ring at the mother–bud junction when tilted at an angle from the glass surface; (b) show a double band at the mother–bud junction; and c show a dot at the mother–bud junction. (C) The fixed cells were double stained with mouse anti-myc and rabbit anti-Cdc11 (septin) primary antibodies and rhodamine-conjugated anti-mouse, and FITC-conjugated anti-rabbit secondary antibodies. (D) Double staining of fixed RLY230 cells with rhodamine phalloidin and anti-myc antibody. Arrows in c point to the actin or Cyk1 band in two large-budded cells. The arrowhead in d points to the actin-free zone in a representative cell with actin patches concentrated around the septum. e shows the ringlike appearance of the actin and Cyk1p containing structure in a cell that was tilted at an angle from the cover glass. Bars, 10 mm.
Figure 5
The Cyk1 ring assembles in anaphase. Fixed RLY230 cells were triple stained with mouse anti-myc, rat antitubulin antibodies, and DAPI. (A) A representative cell that displays a Cyk1 ring and an elongated spindle. (B) Comparison of the intensity of the Cyk1 ring in cells with an elongated spindle (cells a and b) with that in a cell with a short spindle (cell c). The intensities of the Cyk1 rings shown are representative. The ends of the long spindles of cells a and b are out of focus. (C) White bars, cells in an exponentially growing RLY230 population that displayed the spindle and nuclear morphology as diagrammed below the histogram were counted and the resultant numbers were divided by the total number of cells counted (220) to yield the percentages. Solid bars, in the same population, cells that had a Cyk1 ring and the spindle and nuclear morphology as diagrammed were counted. The obtained numbers were divided by the total number of cells with a Cyk1 ring counted (145) to yield the percentages. Bars, 10 μm.
Figure 5
The Cyk1 ring assembles in anaphase. Fixed RLY230 cells were triple stained with mouse anti-myc, rat antitubulin antibodies, and DAPI. (A) A representative cell that displays a Cyk1 ring and an elongated spindle. (B) Comparison of the intensity of the Cyk1 ring in cells with an elongated spindle (cells a and b) with that in a cell with a short spindle (cell c). The intensities of the Cyk1 rings shown are representative. The ends of the long spindles of cells a and b are out of focus. (C) White bars, cells in an exponentially growing RLY230 population that displayed the spindle and nuclear morphology as diagrammed below the histogram were counted and the resultant numbers were divided by the total number of cells counted (220) to yield the percentages. Solid bars, in the same population, cells that had a Cyk1 ring and the spindle and nuclear morphology as diagrammed were counted. The obtained numbers were divided by the total number of cells with a Cyk1 ring counted (145) to yield the percentages. Bars, 10 μm.
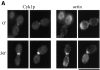
Figure 6
The assembly of the actin ring occurs downstream of Cdc15. RLY238 cells (cdc15-2 CYK1-myc) were grown at 37°C for 3 h. 5 ml of cells were fixed as the 0 time point sample. The rest of the cells were harvested and resuspended in 23°C YPD and grown at 23°C for 15 and 30 min before fixation or processing for immunoblot analysis. The fixed cells were double stained with anti-myc antibody and with rhodamine phalloidin. (A) Representative cells from the 0 time point that had a Cyk1 ring (top left). Representative cells from the 30-min time point that had a Cyk1 ring (bottom left). (B) The number of budded cells (mostly large-budded) that had a Cyk1 ring or an actin ring was counted and divided by the total number of budded cells counted (∼150) to yield the percentage. The number of unbudded cells were also counted and divided by the total number counted to yield the percentage. All cells that had an obvious actin ring had a Cyk1 ring. (C) Cells from each time point were processed for immunoblot analysis using anti-myc antibody. Bar, 10 μm.
Figure 6
The assembly of the actin ring occurs downstream of Cdc15. RLY238 cells (cdc15-2 CYK1-myc) were grown at 37°C for 3 h. 5 ml of cells were fixed as the 0 time point sample. The rest of the cells were harvested and resuspended in 23°C YPD and grown at 23°C for 15 and 30 min before fixation or processing for immunoblot analysis. The fixed cells were double stained with anti-myc antibody and with rhodamine phalloidin. (A) Representative cells from the 0 time point that had a Cyk1 ring (top left). Representative cells from the 30-min time point that had a Cyk1 ring (bottom left). (B) The number of budded cells (mostly large-budded) that had a Cyk1 ring or an actin ring was counted and divided by the total number of budded cells counted (∼150) to yield the percentage. The number of unbudded cells were also counted and divided by the total number counted to yield the percentage. All cells that had an obvious actin ring had a Cyk1 ring. (C) Cells from each time point were processed for immunoblot analysis using anti-myc antibody. Bar, 10 μm.
Figure 6
The assembly of the actin ring occurs downstream of Cdc15. RLY238 cells (cdc15-2 CYK1-myc) were grown at 37°C for 3 h. 5 ml of cells were fixed as the 0 time point sample. The rest of the cells were harvested and resuspended in 23°C YPD and grown at 23°C for 15 and 30 min before fixation or processing for immunoblot analysis. The fixed cells were double stained with anti-myc antibody and with rhodamine phalloidin. (A) Representative cells from the 0 time point that had a Cyk1 ring (top left). Representative cells from the 30-min time point that had a Cyk1 ring (bottom left). (B) The number of budded cells (mostly large-budded) that had a Cyk1 ring or an actin ring was counted and divided by the total number of budded cells counted (∼150) to yield the percentage. The number of unbudded cells were also counted and divided by the total number counted to yield the percentage. All cells that had an obvious actin ring had a Cyk1 ring. (C) Cells from each time point were processed for immunoblot analysis using anti-myc antibody. Bar, 10 μm.
Figure 7
The localization of Myo1p. (A and B) RLY301 (Myo1-myc–expressing) cells were fixed and double stained with mouse anti-myc and rabbit anti-Cdc11 (A) or rhodamine phalloidin (B). (C) RLY302 (Myo1-GFP–expressing) cells were observed by video microscopy as described in Materials and Methods. Arrow in the _0_′ panel indicates the cell discussed in the text. Arrowhead in the _66_′ panel indicates the site where a new Myo1 structure started to appear. Bar, 10 μm.
Figure 8
A model for the pathway of cytokinetic ring assembly in budding yeast cell cycle. Activation of Cdc28 kinase activity by the G1 cyclins (Cln) triggers the assembly of the septin ring and the myosin ring a few minutes before the bud becomes visible. During mitosis, activation of the Cdc15/Cdc5 pathway induces the localization of Cyk1p to the Myo1 ring, which subsequently recruits actin filaments. The septin filaments may disassemble at this point, as suggested by EM studies (Byers and Goetsch, 1976). This could be a trigger for actin ring contraction. After the completion of cytokinesis, the actin ring disassembles immediately by an unknown mechanism. Septum formation then occurs to complete cell division.
Similar articles
- The multiple roles of Cyk1p in the assembly and function of the actomyosin ring in budding yeast.
Shannon KB, Li R. Shannon KB, et al. Mol Biol Cell. 1999 Feb;10(2):283-96. doi: 10.1091/mbc.10.2.283. Mol Biol Cell. 1999. PMID: 9950677 Free PMC article. - Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis.
Bi E, Maddox P, Lew DJ, Salmon ED, McMillan JN, Yeh E, Pringle JR. Bi E, et al. J Cell Biol. 1998 Sep 7;142(5):1301-12. doi: 10.1083/jcb.142.5.1301. J Cell Biol. 1998. PMID: 9732290 Free PMC article. - Saccharomyces cerevisiae Mob1p is required for cytokinesis and mitotic exit.
Luca FC, Mody M, Kurischko C, Roof DM, Giddings TH, Winey M. Luca FC, et al. Mol Cell Biol. 2001 Oct;21(20):6972-83. doi: 10.1128/MCB.21.20.6972-6983.2001. Mol Cell Biol. 2001. PMID: 11564880 Free PMC article. - Cytokinesis in fission yeast: a myosin pas de deux.
Mulvihill DP, Win TZ, Pack TP, Hyams JS. Mulvihill DP, et al. Microsc Res Tech. 2000 Apr 15;49(2):152-60. doi: 10.1002/(SICI)1097-0029(20000415)49:2<152::AID-JEMT7>3.0.CO;2-7. Microsc Res Tech. 2000. PMID: 10816254 Review. - Microtubule and actin-dependent movement of the formin cdc12p in fission yeast.
Chang F. Chang F. Microsc Res Tech. 2000 Apr 15;49(2):161-7. doi: 10.1002/(SICI)1097-0029(20000415)49:2<161::AID-JEMT8>3.0.CO;2-2. Microsc Res Tech. 2000. PMID: 10816255 Review.
Cited by
- Identification of yeast IQGAP (Iqg1p) as an anaphase-promoting-complex substrate and its role in actomyosin-ring-independent cytokinesis.
Ko N, Nishihama R, Tully GH, Ostapenko D, Solomon MJ, Morgan DO, Pringle JR. Ko N, et al. Mol Biol Cell. 2007 Dec;18(12):5139-53. doi: 10.1091/mbc.e07-05-0509. Epub 2007 Oct 17. Mol Biol Cell. 2007. PMID: 17942599 Free PMC article. - Stable and dynamic axes of polarity use distinct formin isoforms in budding yeast.
Pruyne D, Gao L, Bi E, Bretscher A. Pruyne D, et al. Mol Biol Cell. 2004 Nov;15(11):4971-89. doi: 10.1091/mbc.e04-04-0296. Epub 2004 Sep 15. Mol Biol Cell. 2004. PMID: 15371545 Free PMC article. - Septins, under Cla4p regulation, and the chitin ring are required for neck integrity in budding yeast.
Schmidt M, Varma A, Drgon T, Bowers B, Cabib E. Schmidt M, et al. Mol Biol Cell. 2003 May;14(5):2128-41. doi: 10.1091/mbc.e02-08-0547. Epub 2003 Feb 6. Mol Biol Cell. 2003. PMID: 12802080 Free PMC article. - Phosphoinositide signaling plays a key role in cytokinesis.
Janetopoulos C, Devreotes P. Janetopoulos C, et al. J Cell Biol. 2006 Aug 14;174(4):485-90. doi: 10.1083/jcb.200603156. J Cell Biol. 2006. PMID: 16908667 Free PMC article. Review. - Signal transduction pathways regulated by Rho GTPases in Dictyostelium.
Rivero F, Somesh BP. Rivero F, et al. J Muscle Res Cell Motil. 2002;23(7-8):737-49. doi: 10.1023/a:1024423611223. J Muscle Res Cell Motil. 2002. PMID: 12952072 Review.
References
- Berben G, Dumont J, Gilliquet V, Bolle P, Hilger F. The YDp plasmid: a uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. . Yeast. 1991;7:475–477. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases