Murine cytotoxic T lymphocytes recognize an epitope in an EBNA-1 fragment, but fail to lyse EBNA-1-expressing mouse cells - PubMed (original) (raw)
Murine cytotoxic T lymphocytes recognize an epitope in an EBNA-1 fragment, but fail to lyse EBNA-1-expressing mouse cells
S Mukherjee et al. J Exp Med. 1998.
Abstract
Major histocompatibility complex class I-restricted cytotoxic T lymphocytes (CTLs) specific for epitopes within eight of the nine Epstein Barr Virus (EBV)-encoded latency-associated proteins have been recovered from EBV-infected human subjects by restimulation of lymphocytes in vitro. However, human class I-restricted CTL responses capable of recognizing EBNA-1 expressing cells were not detected in these studies. We have raised a murine CTL line that recognizes an epitope within EBNA-1 by immunizing mice with a vaccinia virus encoding a COOH-terminal EBNA-1 fragment. This novel CTL line was used to investigate whether the epitope (positions 509-517 in EBNA-1, presented through Kd) was presented to CTL by mouse cells expressing full-length EBNA-1 or a deletion mutant of EBNA-1, lacking the Glycine-Alanine (Gly-Ala)-rich region. Cells expressing full-length EBNA-1 are not lysed by the CTL line, whereas cells expressing the Gly-Ala deletion mutant are recognized. These results suggest that epitopes from full-length EBNA-1 are poorly presented, and that the Gly-Ala-rich region is responsible for this phenomenon. The inefficient presentation of EBNA-1-derived epitopes may explain the absence or rarity of EBNA-1-specific CTLs in vivo, a strategy that may allow EBV to maintain persistence within the immunocompetent host without being eliminated by CTLs.
Figures
Figure 2
(a) Constructs used to transfect P815 cells. (Top) Full-length EBNA-1 of the B-95-8 sequence (641 amino acids total). Hatched region (93–328) shows the Gly-Ala–rich region. NLS, nuclear localisation sequence as described in reference . The position of the V9L epitope is 509–517 (indicated as V9L). (Bottom) ΔEBNA-1: residues 93–325 have been deleted, but the rest of EBNA-1 is unaffected. (b, i) Western Blot analysis of transfected cells using the Rbt-EBNA-1 antibody (11). Cells are as labeled. A 72-kD band is detected in P815/EBNA-1 cells; based on reactivity with the antiserum and the size of EBNA-1 detected in the 721 LCL line (lane 4), this is EBNA-1. A 43-kD band is detected in P815 cells transfected with EBNA-1 lacking the Gly-Ala repeat. (b, ii) Western Blot using the P-107 serum (12) that reacts against the Gly-Ala repeat only. Lanes 2 and 3 indicate P815 cells transfected with vectors alone (i.e., no EBNA-1). Lane 6, NAD20 cells are EBV-transformed LCLs. (c) Cell staining with Rbt-EBNA-1 serum (11) reactive against EBNA-1. Nuclear staining is indicated with N and arrow. Cells are as indicated in legend and text.
Figure 2
(a) Constructs used to transfect P815 cells. (Top) Full-length EBNA-1 of the B-95-8 sequence (641 amino acids total). Hatched region (93–328) shows the Gly-Ala–rich region. NLS, nuclear localisation sequence as described in reference . The position of the V9L epitope is 509–517 (indicated as V9L). (Bottom) ΔEBNA-1: residues 93–325 have been deleted, but the rest of EBNA-1 is unaffected. (b, i) Western Blot analysis of transfected cells using the Rbt-EBNA-1 antibody (11). Cells are as labeled. A 72-kD band is detected in P815/EBNA-1 cells; based on reactivity with the antiserum and the size of EBNA-1 detected in the 721 LCL line (lane 4), this is EBNA-1. A 43-kD band is detected in P815 cells transfected with EBNA-1 lacking the Gly-Ala repeat. (b, ii) Western Blot using the P-107 serum (12) that reacts against the Gly-Ala repeat only. Lanes 2 and 3 indicate P815 cells transfected with vectors alone (i.e., no EBNA-1). Lane 6, NAD20 cells are EBV-transformed LCLs. (c) Cell staining with Rbt-EBNA-1 serum (11) reactive against EBNA-1. Nuclear staining is indicated with N and arrow. Cells are as indicated in legend and text.
Figure 2
(a) Constructs used to transfect P815 cells. (Top) Full-length EBNA-1 of the B-95-8 sequence (641 amino acids total). Hatched region (93–328) shows the Gly-Ala–rich region. NLS, nuclear localisation sequence as described in reference . The position of the V9L epitope is 509–517 (indicated as V9L). (Bottom) ΔEBNA-1: residues 93–325 have been deleted, but the rest of EBNA-1 is unaffected. (b, i) Western Blot analysis of transfected cells using the Rbt-EBNA-1 antibody (11). Cells are as labeled. A 72-kD band is detected in P815/EBNA-1 cells; based on reactivity with the antiserum and the size of EBNA-1 detected in the 721 LCL line (lane 4), this is EBNA-1. A 43-kD band is detected in P815 cells transfected with EBNA-1 lacking the Gly-Ala repeat. (b, ii) Western Blot using the P-107 serum (12) that reacts against the Gly-Ala repeat only. Lanes 2 and 3 indicate P815 cells transfected with vectors alone (i.e., no EBNA-1). Lane 6, NAD20 cells are EBV-transformed LCLs. (c) Cell staining with Rbt-EBNA-1 serum (11) reactive against EBNA-1. Nuclear staining is indicated with N and arrow. Cells are as indicated in legend and text.
Figure 2
(a) Constructs used to transfect P815 cells. (Top) Full-length EBNA-1 of the B-95-8 sequence (641 amino acids total). Hatched region (93–328) shows the Gly-Ala–rich region. NLS, nuclear localisation sequence as described in reference . The position of the V9L epitope is 509–517 (indicated as V9L). (Bottom) ΔEBNA-1: residues 93–325 have been deleted, but the rest of EBNA-1 is unaffected. (b, i) Western Blot analysis of transfected cells using the Rbt-EBNA-1 antibody (11). Cells are as labeled. A 72-kD band is detected in P815/EBNA-1 cells; based on reactivity with the antiserum and the size of EBNA-1 detected in the 721 LCL line (lane 4), this is EBNA-1. A 43-kD band is detected in P815 cells transfected with EBNA-1 lacking the Gly-Ala repeat. (b, ii) Western Blot using the P-107 serum (12) that reacts against the Gly-Ala repeat only. Lanes 2 and 3 indicate P815 cells transfected with vectors alone (i.e., no EBNA-1). Lane 6, NAD20 cells are EBV-transformed LCLs. (c) Cell staining with Rbt-EBNA-1 serum (11) reactive against EBNA-1. Nuclear staining is indicated with N and arrow. Cells are as indicated in legend and text.
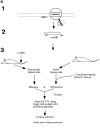
Figure 1
(a) Scheme for rescue of peptide-specific CTLs by coculturing immunized splenocytes with naive splenocytes pulsed with candidate peptides (see text). (1) Identification of candidate epitopes. (2) Construction of vaccinia virus expressing the EBNA-1 (505–583) fragment (called VVΔEB-1) containing the candidate epitopes. (3) Immunization of mice and recovery of CTLs. (b) Mouse strains initially immunized with VVΔEB-1 and the peptides used to restimulate them in vitro. (Column 1) Candidate epitopes derived from the COOH-terminal of EBNA-1 using consensus motifs described in reference . (Column 2) Class I molecule for which the candidate peptide in column 1 carries the consensus motif (and strains immunized with VVΔEB-1). For example, V
Y
GGSKTS
L
contains a Kd motif (i.e., Y at position 2, and L at position 9, underlined). Splenocytes from BALB/c mice (H2-d) immunized with VVΔEB-1 were restimulated with splenocytes pulsed with the V9L peptide, as described in a. Cocultures restimulated on peptides (as in column 1) were used as effectors in a Cr–release assay using peptide-pulsed cells as targets (see below for the target cells). (Column 3) Specific lysis above background in 51Cr release assays 5 d after in vitro restimulation with peptide-pulsed cells, was obtained for each of the effector cultures. Target cells used: P815 for all H2-d, L929 for H2-k, and EL-4 for H2-b. (c) V9L CTLs are peptide specific and MHC restricted. P815 cells were pulsed with the peptide dose indicated (in nM) and used as targets in Cr–release assay. K/T ratio was 20:1. Unpulsed cells (0 nM) are not lysed. Hatched bar shows L/Db cells that were pulsed with V9L peptide; these cells do not express Kd and were not lysed. The x-axis represents P815 cells pulsed with peptide at concentration indicated (in nM) and used as target for V9L CTLs.
Figure 1
(a) Scheme for rescue of peptide-specific CTLs by coculturing immunized splenocytes with naive splenocytes pulsed with candidate peptides (see text). (1) Identification of candidate epitopes. (2) Construction of vaccinia virus expressing the EBNA-1 (505–583) fragment (called VVΔEB-1) containing the candidate epitopes. (3) Immunization of mice and recovery of CTLs. (b) Mouse strains initially immunized with VVΔEB-1 and the peptides used to restimulate them in vitro. (Column 1) Candidate epitopes derived from the COOH-terminal of EBNA-1 using consensus motifs described in reference . (Column 2) Class I molecule for which the candidate peptide in column 1 carries the consensus motif (and strains immunized with VVΔEB-1). For example, V
Y
GGSKTS
L
contains a Kd motif (i.e., Y at position 2, and L at position 9, underlined). Splenocytes from BALB/c mice (H2-d) immunized with VVΔEB-1 were restimulated with splenocytes pulsed with the V9L peptide, as described in a. Cocultures restimulated on peptides (as in column 1) were used as effectors in a Cr–release assay using peptide-pulsed cells as targets (see below for the target cells). (Column 3) Specific lysis above background in 51Cr release assays 5 d after in vitro restimulation with peptide-pulsed cells, was obtained for each of the effector cultures. Target cells used: P815 for all H2-d, L929 for H2-k, and EL-4 for H2-b. (c) V9L CTLs are peptide specific and MHC restricted. P815 cells were pulsed with the peptide dose indicated (in nM) and used as targets in Cr–release assay. K/T ratio was 20:1. Unpulsed cells (0 nM) are not lysed. Hatched bar shows L/Db cells that were pulsed with V9L peptide; these cells do not express Kd and were not lysed. The x-axis represents P815 cells pulsed with peptide at concentration indicated (in nM) and used as target for V9L CTLs.
Figure 1
(a) Scheme for rescue of peptide-specific CTLs by coculturing immunized splenocytes with naive splenocytes pulsed with candidate peptides (see text). (1) Identification of candidate epitopes. (2) Construction of vaccinia virus expressing the EBNA-1 (505–583) fragment (called VVΔEB-1) containing the candidate epitopes. (3) Immunization of mice and recovery of CTLs. (b) Mouse strains initially immunized with VVΔEB-1 and the peptides used to restimulate them in vitro. (Column 1) Candidate epitopes derived from the COOH-terminal of EBNA-1 using consensus motifs described in reference . (Column 2) Class I molecule for which the candidate peptide in column 1 carries the consensus motif (and strains immunized with VVΔEB-1). For example, V
Y
GGSKTS
L
contains a Kd motif (i.e., Y at position 2, and L at position 9, underlined). Splenocytes from BALB/c mice (H2-d) immunized with VVΔEB-1 were restimulated with splenocytes pulsed with the V9L peptide, as described in a. Cocultures restimulated on peptides (as in column 1) were used as effectors in a Cr–release assay using peptide-pulsed cells as targets (see below for the target cells). (Column 3) Specific lysis above background in 51Cr release assays 5 d after in vitro restimulation with peptide-pulsed cells, was obtained for each of the effector cultures. Target cells used: P815 for all H2-d, L929 for H2-k, and EL-4 for H2-b. (c) V9L CTLs are peptide specific and MHC restricted. P815 cells were pulsed with the peptide dose indicated (in nM) and used as targets in Cr–release assay. K/T ratio was 20:1. Unpulsed cells (0 nM) are not lysed. Hatched bar shows L/Db cells that were pulsed with V9L peptide; these cells do not express Kd and were not lysed. The x-axis represents P815 cells pulsed with peptide at concentration indicated (in nM) and used as target for V9L CTLs.
Figure 3
V9L CTLs used as effectors in 51Cr–release assays. Targets: P815/EBNA-1 (open circles), P815/ΔEBNA-1 (filled squares), P815 with no peptide (open triangles), and a P815 + V9L peptide pulsed at 10 μm (filled circles).
Similar articles
- Localization of Epstein-Barr virus cytotoxic T cell epitopes using recombinant vaccinia: implications for vaccine development.
Khanna R, Burrows SR, Kurilla MG, Jacob CA, Misko IS, Sculley TB, Kieff E, Moss DJ. Khanna R, et al. J Exp Med. 1992 Jul 1;176(1):169-76. doi: 10.1084/jem.176.1.169. J Exp Med. 1992. PMID: 1377222 Free PMC article. - Class I major histocompatibility complex-restricted cytotoxic T lymphocytes specific for Epstein-Barr virus (EBV)-transformed B lymphoblastoid cell lines against which they were raised.
Hill AB, Lee SP, Haurum JS, Murray N, Yao QY, Rowe M, Signoret N, Rickinson AB, McMichael AJ. Hill AB, et al. J Exp Med. 1995 Jun 1;181(6):2221-8. doi: 10.1084/jem.181.6.2221. J Exp Med. 1995. PMID: 7539044 Free PMC article. - T cell recognition of Epstein-Barr virus associated lymphomas.
Rickinson AB, Murray RJ, Brooks J, Griffin H, Moss DJ, Masucci MG. Rickinson AB, et al. Cancer Surv. 1992;13:53-80. Cancer Surv. 1992. PMID: 1330300 Review. - HIV epitopes recognized by cytotoxic T-lymphocytes.
Autran B, Letvin NL. Autran B, et al. AIDS. 1991;5 Suppl 2:S145-50. doi: 10.1097/00002030-199101001-00020. AIDS. 1991. PMID: 1726954 Review. No abstract available.
Cited by
- Inhibition of antigen presentation by the glycine/alanine repeat domain is not conserved in simian homologues of Epstein-Barr virus nuclear antigen 1.
Blake NW, Moghaddam A, Rao P, Kaur A, Glickman R, Cho YG, Marchini A, Haigh T, Johnson RP, Rickinson AB, Wang F. Blake NW, et al. J Virol. 1999 Sep;73(9):7381-9. doi: 10.1128/JVI.73.9.7381-7389.1999. J Virol. 1999. PMID: 10438828 Free PMC article. - Endogenous presentation of CD8+ T cell epitopes from Epstein-Barr virus-encoded nuclear antigen 1.
Tellam J, Connolly G, Green KJ, Miles JJ, Moss DJ, Burrows SR, Khanna R. Tellam J, et al. J Exp Med. 2004 May 17;199(10):1421-31. doi: 10.1084/jem.20040191. J Exp Med. 2004. PMID: 15148340 Free PMC article. - Inhibition of proteasomal degradation by the gly-Ala repeat of Epstein-Barr virus is influenced by the length of the repeat and the strength of the degradation signal.
Dantuma NP, Heessen S, Lindsten K, Jellne M, Masucci MG. Dantuma NP, et al. Proc Natl Acad Sci U S A. 2000 Jul 18;97(15):8381-5. doi: 10.1073/pnas.140217397. Proc Natl Acad Sci U S A. 2000. PMID: 10890896 Free PMC article. - CD8 T cell recognition of endogenously expressed epstein-barr virus nuclear antigen 1.
Lee SP, Brooks JM, Al-Jarrah H, Thomas WA, Haigh TA, Taylor GS, Humme S, Schepers A, Hammerschmidt W, Yates JL, Rickinson AB, Blake NW. Lee SP, et al. J Exp Med. 2004 May 17;199(10):1409-20. doi: 10.1084/jem.20040121. J Exp Med. 2004. PMID: 15148339 Free PMC article. - Gamma-herpesvirus latency requires T cell evasion during episome maintenance.
Bennett NJ, May JS, Stevenson PG. Bennett NJ, et al. PLoS Biol. 2005 Apr;3(4):e120. doi: 10.1371/journal.pbio.0030120. Epub 2005 Mar 22. PLoS Biol. 2005. PMID: 15769185 Free PMC article.
References
- Kieff, E. 1996. Epstein Barr virus and its replication. In Virology. B.N. Fields, D.M. Knipe, P. Howley, R. Chanock, J. Melnick, T. Monath, B. Rozman, S.E. Straus, editors. Lippincott-Raven Press, New York. 2243–2396.
- Klein G. Viral latency and transformation: the strategy of Epstein Barr virus. Cell. 1989;58:5–8. - PubMed
- Townsend A, Bodmer H. Antigen recognition by class I–restricted T lymphocytes. Annu Rev Immunol. 1989;7:601–624. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases