The myogenic regulatory gene Mef2 is a direct target for transcriptional activation by Twist during Drosophila myogenesis - PubMed (original) (raw)
The myogenic regulatory gene Mef2 is a direct target for transcriptional activation by Twist during Drosophila myogenesis
R M Cripps et al. Genes Dev. 1998.
Abstract
MEF2 is a MADS-box transcription factor required for muscle development in Drosophila. Here, we show that the bHLH transcription factor Twist directly regulates Mef2 expression in adult somatic muscle precursor cells via a 175-bp enhancer located 2245 bp upstream of the transcriptional start site. Within this element, a single evolutionarily conserved E box is essential for enhancer activity. Twist protein can bind to this E box to activate Mef2 transcription, and ectopic expression of twist results in ectopic activation of the wild-type 175-bp enhancer. By use of a temperature-sensitive mutant of twist, we show that activation of Mef2 transcription via this enhancer by Twist is required for normal adult muscle development, and reduction in Twist function results in phenotypes similar to those observed previously in Mef2 mutant adults. The 175-bp enhancer is also active in the embryonic mesoderm, indicating that this enhancer functions at multiple times during development, and its function is dependent on the same conserved E box. In embryos, a reduction in Twist function also strongly reduced Mef2 expression. These findings define a novel transcriptional pathway required for skeletal muscle development and identify Twist as an essential and direct regulator of Mef2 expression in the somatic mesoderm.
Figures
Figure 1
Characterization of the Mef2 adepithelial cell enhancer. (A) The top line shows the genomic organization of the D. melanogaster Mef2 locus. Solid and open boxes represent coding and noncoding transcribed regions, respectively. Vertical ticks labeled R show the locations of _Eco_RI restriction sites (R in parentheses represents a synthetic site; see Schulz et al. 1996). Each of these _Eco_RI fragments, cloned upstream of the lacZ gene, was tested for expression in the wing disc adepithelial cells. Only the fragment from −3564 to +521 showed expression. Below is an expansion of this ∼4-kb region, within which the indicated fragments were tested. Summaries of their expression in wing imaginal discs are shown at right. The shaded bar denotes a 185-bp region that is required for expression in adepithelial cells. (B) Sequence similarity between D. melanogaster and D. virilis in the 185-bp region. Strong similarity is observed over a 100-bp segment. The D. virilis sequence shown is from −2448 to −2359. (Dm) D. melanogaster sequence; (Dv) D. virilis sequence. Three E boxes are underlined, and E1 and E2 are indicated.
Figure 2
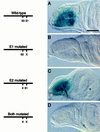
Expression of the 175-bp adepithelial cell enhancer in wing imaginal discs depends on the E1 E box. Transgenic lines carrying enhancer–lacZ constructs were tested for β-galactosidase expression in wing imaginal discs. (A) Wild-type 175-bp enhancer drives high levels of expression in the adepithelial cells; (B) enhancer with E1 mutated has no expression; (C) enhancer with E2 mutated drives lacZ expression at wild-type levels; (D) the enhancer with both E1 and E2 mutated has no activity. Bar, 50 μm.
Figure 3
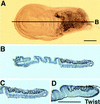
Mef2 and twist are expressed in overlapping domains in wing imaginal discs. Third larval instar wing imaginal discs were reacted with either anti-MEF2 (A) or anti-Twist (not shown) polyclonal antibodies. Discs were then embedded in paraffin and sectioned to localize the immunoreactive cells. Anti-MEF2 staining was localized to the proximal end of the disc (B), and closer observation (C) revealed that it was expressed in most or all of the cells in this region. Twist protein accumulated in a very similar pattern (D), showing that these two proteins are predominantly coexpressed in the adult muscle precursor cells. Bar, 50 μm.
Figure 4
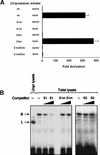
Twist activates the 175-bp enhancer in Drosophila SL2 cells. (A) SL2 cells were transfected with the indicated β-galactosidase reporter plasmids and either twist expression vector or expression vector without a cDNA insert. The data represent the mean values obtained in four independent transfections and are expressed as the fold-activation obtained in each sample over the activity generated by the wild-type enhancer construct in the absence of twist expression plasmid. Error bars represent the standard error of the means for the four experiments. The mean level of β-galactosidase activity produced by the wild-type enhancer construct in the absence of Twist was 3.4 × 10−6 units and in the presence of Twist was 1.1 × 10−3 units (one unit is defined as the amount of enzyme that will convert >95% of the β-
d
-galactose from 1 nmole of Galβ1–3GlcNAcβ1–3Galβ1–4Glc–AMC in a 10-μl reaction in 1 hr at 37°C and at pH 4.5). (B) Twist binding to the E1 E box. An oligonucleotide corresponding to the E1 E box was used as a probe in an electrophoretic mobility shift assay by use of in vitro-translated Twist. Unprogrammed lysate was included in a parallel lane. A 50- or 300-fold excess of unlabeled E1, unlabeled E1 mutant (E1m), or unlabeled E2 oligonucleotides were used as competitors. Competition by the wild-type E1 oligonucleotide but neither the E1 mutant nor E2 indicates a specific interaction between Twist and E1. B, probe bound with Twist; L, complex detected in unprogrammed lysate.
Figure 5
Twist activates the 175-bp enhancer in embryos. Embryos carrying either the wild-type 175-bp enhancer–lacZ construct (A,C) or the E1 mutated construct (B,D) were stained for β-galactosidase protein in the absence (A,B) or presence (C,D) of ectopically expressed twist. The 175-bp enhancer was active in wild-type embryos (A; see also Fig. 11), and activity was induced throughout the embryo after ubiquitous expression of twist (C). The E1 mutated enhancer was inactive in wild-type embryos (B) and was not induced by ectopic Twist (D), although the morphology of this embryo was affected. Bar, 100 μm.
Figure 6
Expression of adepithelial cell markers in wild-type and RY50/V50 twist mutants. (A) (Left) Wild type. (Right) RY50/V50 raised at the restrictive temperature. (Top) Immunostaining by an anti-MEF2 polyclonal antibody. (Center) In situ hybridization with a digoxigenin-labeled antisense probe for the Drosophila fibroblast growth factor receptor homolog -1, htl. (Bottom) Immunostaining by an anti-Cut monoclonal antibody. Note that MEF2 accumulation is greatly reduced, yet the other markers are expressed at normal levels. Bar, 50 μm. (B) A ∼3.4-kb Mef2 enhancer/promoter fragment is sensitive to the levels of twist expression. Transgenic flies carrying the −2884 to +521 fragment cloned upstream of lacZ were crossed into the twist mutant backgrounds. RY50/V50 mutants raised at either the permissive (left, 18°C) or restrictive (right, 30°C) temperatures were analyzed for β-galactosidase activity in wing imaginal discs. Transgene expression was only observed at 18°C, indicating that Twist function is required for enhancer activity. Bar, 25 μm.
Figure 7
Adult muscle phenotypes resulting from reduced twist expression. (A–C) vertical transverse paraffin sections of thoraces to visualize the two rows of dorsal longitudinal indirect flight muscles (muscles are numbered 1–6 where present). (A) wild-type; (B) RY50/V50 raised at 18°C; (C) RY50/V50 raised at 30°C. Bar, 100 μm. (D–F) summary of temperature shifts performed and fiber defects observed; fiber numbers represent the average number of DLM fibers per hemithorax. (D) RY50/V50 raised at 30°C after embryogenesis; (E) RY50/V50 raised at 18°C; (F) wild-type raised at 30°C after embryogenesis. (
s.e.m.
) Standard error of the mean. Developmental stages: (E) embryonic stage; (L1–3) larval instars; (P) pupal stage; (A) adult stage.
Figure 8
Expression of the 175-bp enhancer during embryogenesis. (A) At stage 9, the enhancer was only weakly active in the developing mesoderm and was segmentally modulated. (B) at late stage 11 the enhancer was active strongly in segmentally repeating groups of cells that are the precursors of the larval somatic muscles (arrows). (C) By stage 15, enhancer activity was waning. (D–E) Accumulation of MEF2 in the presence (D) or absence (E) of Twist function after gastrulation. (D) MEF2 acumulation in a late stage 11 twist mutant heterozygous embryo, showing normal expression in somatic muscle myoblasts (arrows). (E) MEF2 accumulation in a sibling temperature-sensitive twist mutant in which Twist function had been reduced after gastrulation. There is a general reduction in Mef2 expression in the somatic myoblasts (arrows), as well as in the precursors of the pharyngeal muscles (out of focus at anterior end). Bar, 100 μm.
Similar articles
- Combinatorial control of Drosophila mef2 gene expression in cardiac and somatic muscle cell lineages.
Gajewski K, Kim Y, Choi CY, Schulz RA. Gajewski K, et al. Dev Genes Evol. 1998 Sep;208(7):382-92. doi: 10.1007/s004270050194. Dev Genes Evol. 1998. PMID: 9732552 - Myocyte-specific enhancer factor 2 acts cooperatively with a muscle activator region to regulate Drosophila tropomyosin gene muscle expression.
Lin MH, Nguyen HT, Dybala C, Storti RV. Lin MH, et al. Proc Natl Acad Sci U S A. 1996 May 14;93(10):4623-8. doi: 10.1073/pnas.93.10.4623. Proc Natl Acad Sci U S A. 1996. PMID: 8643453 Free PMC article. - A series of mutations in the D-MEF2 transcription factor reveal multiple functions in larval and adult myogenesis in Drosophila.
Ranganayakulu G, Zhao B, Dokidis A, Molkentin JD, Olson EN, Schulz RA. Ranganayakulu G, et al. Dev Biol. 1995 Sep;171(1):169-81. doi: 10.1006/dbio.1995.1269. Dev Biol. 1995. PMID: 7556894 - Muscle development: a transcriptional pathway in myogenesis.
Taylor MV. Taylor MV. Curr Biol. 1998 May 7;8(10):R356-8. doi: 10.1016/s0960-9822(98)70221-0. Curr Biol. 1998. PMID: 9601637 Review. - Muscle development. Making Drosophila muscle.
Taylor MV. Taylor MV. Curr Biol. 1995 Jul 1;5(7):740-2. doi: 10.1016/s0960-9822(95)00149-7. Curr Biol. 1995. PMID: 7583119 Review.
Cited by
- Analysis of skeletal muscle development in Drosophila.
Morriss GR, Bryantsev AL, Chechenova M, LaBeau EM, Lovato TL, Ryan KM, Cripps RM. Morriss GR, et al. Methods Mol Biol. 2012;798:127-52. doi: 10.1007/978-1-61779-343-1_8. Methods Mol Biol. 2012. PMID: 22130835 Free PMC article. - Combinatorial binding predicts spatio-temporal cis-regulatory activity.
Zinzen RP, Girardot C, Gagneur J, Braun M, Furlong EE. Zinzen RP, et al. Nature. 2009 Nov 5;462(7269):65-70. doi: 10.1038/nature08531. Nature. 2009. PMID: 19890324 - Drosophila as a Genetic Model for Hematopoiesis.
Banerjee U, Girard JR, Goins LM, Spratford CM. Banerjee U, et al. Genetics. 2019 Feb;211(2):367-417. doi: 10.1534/genetics.118.300223. Genetics. 2019. PMID: 30733377 Free PMC article. Review. - Targeted inactivation of the rickets receptor in muscle compromises Drosophila viability.
Harwood BN, Draper I, Kopin AS. Harwood BN, et al. J Exp Biol. 2014 Nov 15;217(Pt 22):4091-8. doi: 10.1242/jeb.110098. Epub 2014 Oct 2. J Exp Biol. 2014. PMID: 25278473 Free PMC article. - Differential regulation of mesodermal gene expression by Drosophila cell type-specific Forkhead transcription factors.
Zhu X, Ahmad SM, Aboukhalil A, Busser BW, Kim Y, Tansey TR, Haimovich A, Jeffries N, Bulyk ML, Michelson AM. Zhu X, et al. Development. 2012 Apr;139(8):1457-66. doi: 10.1242/dev.069005. Epub 2012 Feb 29. Development. 2012. PMID: 22378636 Free PMC article.
References
- Ashburner M. Drosophila: A laboratory handbook and manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989.
- Bate M. The mesoderm and its derivatives. In: Bate M, Martinez Arias A, editors. The development of Drosophila melanogaster. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 1013–1090.
- Bate M, Rushton E, Currie DA. Cells with persistent twist expression are the embryonic precursors of adult muscles in Drosophila. Development. 1991;113:79–89. - PubMed
- Baylies MK, Bate M. twist: A myogenic switch in Drosophila. Science. 1996;272:1481–1484. - PubMed
- Bernstein SI, O’Donnell PT, Cripps RM. Molecular genetic analysis of muscle development, structure and function in Drosophila. Int Rev Cytol. 1993;43:63–152. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases