Recognition signal for C-mannosylation of Trp-7 in RNase 2 consists of sequence Trp-x-x-Trp - PubMed (original) (raw)
Recognition signal for C-mannosylation of Trp-7 in RNase 2 consists of sequence Trp-x-x-Trp
J Krieg et al. Mol Biol Cell. 1998 Feb.
Free PMC article
Abstract
C2-alpha-Mannosyltryptophan was discovered in human RNase 2, an enzyme that occurs in eosinophils and is involved in host defense. It represents a novel way of attaching carbohydrate to a protein in addition to the well-known N- and O-glycosylations. The reaction is specific, as in RNase 2 Trp-7, but never Trp-10, which is modified. In this article, we address which structural features provide the specificity of the reaction. Expression of chimeras of RNase 2 and nonglycosylated RNase 4 and deletion mutants in HEK293 cells identified residues 1-13 to be sufficient for C-mannosylation. Site-directed mutagenesis revealed the sequence Trp-x-x-Trp, in which the first Trp becomes mannosylated, as the specificity determinant. The Trp residue at position +3 can be replaced by Phe, which reduces the efficiency of the reaction threefold. Interpretation of the data in the context of the three-dimensional structure of RNase 2 strongly suggests that the primary, rather than the tertiary, structure forms the determinant. The sequence motif occurs in 336 mammalian proteins currently present in protein databases. Two of these proteins were analyzed protein chemically, which showed partial C-glycosylation of recombinant human interleukin 12. The frequent occurrence of the protein recognition motif suggests that C-glycosides could be part of the structure of more proteins than assumed so far.
Figures
Figure 1
Structure of hybrid RNases. (A) The RNase 2 portion of a hybrid is depicted as an open rectangle, whereas that of RNase 4 (numbering in italic) is hatched. Trp-7 has been underlined. (B) Comparison of the primary structures of human RNase 2 and porcine RNase 4. Amino acids in common are indicated by a line when they occur at the surface of the protein and by a colon when they are buried.
Figure 2
Reversed-phase HPLC purification of RNase 2.4. Immunopurified RNase 2.4 was chromatographed on a C4 column (1 mm in diameter) equilibrated in 0.1% TFA. A linear gradient of 0–80% solvent B (70% CH3CN in 0.085% TFA) over 75 min was used at a flow rate of 50 μl/min. The inset shows a Western blot analysis of the fractions using antibodies against RNase 4 or modification-specific antibodies, α(5–10).
Figure 3
Western blot analysis of single-site mutants of RNase 2.4. Approximately 0.5 μg of each protein was electrophoresed on a 15% SDS-PAA gel and blotted onto nitrocellulose. The blots were probed with modification-specific antibodies, α(5–10), and, after stripping, with αRNase 4 antibodies.
Figure 4
Characterization of RNase 2.4 and its mutants. (A) Five micrograms of reduced and carboxymethylated RNase were digested at glutamic acid residues with endoproteinase Glu-C and fractionated by C8 reversed-phase LC-ESIMS. A 1-mm diameter column equilibrated in 95% solvent A (2% CH3CN, 0.05% TFA) and 5% solvent B (80% CH3CN, 0.045% TFA) was used. Peptides were eluted with a linear gradient of 5–40% solvent B at a flow rate of 50 μl/min. C-mannosylated (“m”) and unmodified (“u”) fragment −4 to 12 were assigned based on their molecular masses. The results obtained with wild-type RNase 2.4 (upper panel), mutant T6A (middle panel), and W10A (lower panel) are shown as representative examples. (B) Peptide −4 to 12 was digested with elastase and fractionated by reversed-phase LC-ESIMS. The peptide map obtained from the T6A mutant is shown as a representative example. Peptide 9–12 with unmodified Trp-10, 608 Da; peptide −4 to 8 with C-mannosylated Trp-7, 1542 Da. (C) ESIMS of the fragment −4 to 12 from RNase 2.4, W10A. The molecular mass of 1885 Da corresponds to that of the peptide with unmodified Trp.
Figure 5
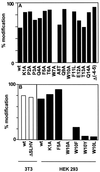
Mutational analysis of RNase 2.4 and RNase 2. (A) The degree of C-mannosylation of Trp-7 in the indicated mutants of hybrid RNase 2.4 was determined from the ratio of modified and unmodified fragments −4 to 12 and is depicted by filled bars. (B) The degree of modification of Trp-7 in the indicated mutants of RNase 2 was determined as in A and are plotted as open bars for the experiment performed in 3T3 cells and as filled bars for the experiment performed in HEK293 cells. The data represent the average of at least two independent experiments. The SD was 1–16% of the mean.
Figure 6
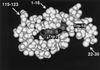
Three-dimensional structure around the C-mannosylation site of recombinant RNase 2. The indole moieties of Trp-7 and -10 are shown in dark gray. Since the protein was produced in E. coli, Trp-7 is not C-mannosylated. The coordinates used to produce this figure were obtained from Mosimann et al., 1996.
Similar articles
- Spectroscopic and protein chemical analyses demonstrate the presence of C-mannosylated tryptophan in intact human RNase 2 and its isoforms.
Löffler A, Doucey MA, Jansson AM, Müller DR, de Beer T, Hess D, Meldal M, Richter WJ, Vliegenthart JF, Hofsteenge J. Löffler A, et al. Biochemistry. 1996 Sep 17;35(37):12005-14. doi: 10.1021/bi9610515. Biochemistry. 1996. PMID: 8810905 - C-Mannosylation of human RNase 2 is an intracellular process performed by a variety of cultured cells.
Krieg J, Gläsner W, Vicentini A, Doucey MA, Löffler A, Hess D, Hofsteenge J. Krieg J, et al. J Biol Chem. 1997 Oct 17;272(42):26687-92. doi: 10.1074/jbc.272.42.26687. J Biol Chem. 1997. PMID: 9334252 - Protein C-mannosylation is enzyme-catalysed and uses dolichyl-phosphate-mannose as a precursor.
Doucey MA, Hess D, Cacan R, Hofsteenge J. Doucey MA, et al. Mol Biol Cell. 1998 Feb;9(2):291-300. doi: 10.1091/mbc.9.2.291. Mol Biol Cell. 1998. PMID: 9450955 Free PMC article. - Protein _C_-Mannosylation and _C_-Mannosyl Tryptophan in Chemical Biology and Medicine.
Minakata S, Manabe S, Inai Y, Ikezaki M, Nishitsuji K, Ito Y, Ihara Y. Minakata S, et al. Molecules. 2021 Aug 30;26(17):5258. doi: 10.3390/molecules26175258. Molecules. 2021. PMID: 34500691 Free PMC article. Review. - Protein C-mannosylation: facts and questions.
Furmanek A, Hofsteenge J. Furmanek A, et al. Acta Biochim Pol. 2000;47(3):781-9. Acta Biochim Pol. 2000. PMID: 11310977 Review.
Cited by
- Re-mining serum proteomics data reveals extensive post-translational modifications upon Zika and dengue infection.
Allgoewer K, Wu S, Choi H, Vogel C. Allgoewer K, et al. Mol Omics. 2023 May 9;19(4):308-320. doi: 10.1039/d2mo00258b. Mol Omics. 2023. PMID: 36810580 Free PMC article. - Ebola sGP--the first viral glycoprotein shown to be C-mannosylated.
Falzarano D, Krokhin O, Van Domselaar G, Wolf K, Seebach J, Schnittler HJ, Feldmann H. Falzarano D, et al. Virology. 2007 Nov 10;368(1):83-90. doi: 10.1016/j.virol.2007.06.015. Epub 2007 Jul 20. Virology. 2007. PMID: 17659315 Free PMC article. - Ridge regression estimated linear probability model predictions of O-glycosylation in proteins with structural and sequence data.
Gana R, Vasudevan S. Gana R, et al. BMC Mol Cell Biol. 2019 Jun 28;20(1):21. doi: 10.1186/s12860-019-0200-9. BMC Mol Cell Biol. 2019. PMID: 31253080 Free PMC article. - HDAC-6 interacts with and deacetylates tubulin and microtubules in vivo.
Zhang Y, Li N, Caron C, Matthias G, Hess D, Khochbin S, Matthias P. Zhang Y, et al. EMBO J. 2003 Mar 3;22(5):1168-79. doi: 10.1093/emboj/cdg115. EMBO J. 2003. PMID: 12606581 Free PMC article. - Involvement of DPY19L3 in Myogenic Differentiation of C2C12 Myoblasts.
Mori K, Sun H, Miura K, Simizu S. Mori K, et al. Molecules. 2021 Sep 19;26(18):5685. doi: 10.3390/molecules26185685. Molecules. 2021. PMID: 34577156 Free PMC article.
References
- Baranski TJ, Faust PL, Kornfeld S. Generation of a lysosomal targeting signal in the secretory protein pepsinogen. Cell. 1990;63:281–291. - PubMed
- Baranski TJ, Koelsch G, Hartsuck JA, Kornfeld S. Mapping and molecular modeling of a recognition domain for lysosomal enzyme targeting. J Biol Chem. 1991;266:23365–23372. - PubMed
- Beintema JJ, Hofsteenge J, Iwama M, Morita T, Ohgi K, Irie M, Sugiyama RH, Schieven GL, Dekker CA, Glitz DG. Amino acid sequence of the nonsecretory ribonuclease of human urine. Biochemistry. 1988;27:4530–4538. - PubMed
- Bergwerff AA, Oostrum J, Asselbergs AM, Buergi R, Hokke H, Kamerling JP, Vliegenthart FG. Primary structure of N-linked carbohydrate chains of a human chimeric plasminogen activator K2tu-PA expressed in Chinese hamster ovary cells. Eur J Biochem. 1993;212:639–656. - PubMed
- de Beer T, Vliegenthart JFG, Löffler A, Hofsteenge J. The hexopyranosyl residue that is C-glycosidically linked to the side chain of tryptophan-7 in human RNase Us is α-mannopyranose. Biochemistry. 1995;34:11785–11789. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous