SUMO-1 modification and its role in targeting the Ran GTPase-activating protein, RanGAP1, to the nuclear pore complex - PubMed (original) (raw)
SUMO-1 modification and its role in targeting the Ran GTPase-activating protein, RanGAP1, to the nuclear pore complex
M J Matunis et al. J Cell Biol. 1998.
Abstract
RanGAP1 is the GTPase-activating protein for Ran, a small ras-like GTPase involved in regulating nucleocytoplasmic transport. In vertebrates, RanGAP1 is present in two forms: one that is cytoplasmic, and another that is concentrated at the cytoplasmic fibers of nuclear pore complexes (NPCs). The NPC-associated form of RanGAP1 is covalently modified by the small ubiquitin-like protein, SUMO-1, and we have recently proposed that SUMO-1 modification functions to target RanGAP1 to the NPC. Here, we identify the domain of RanGAP1 that specifies SUMO-1 modification and demonstrate that mutations in this domain that inhibit modification also inhibit targeting to the NPC. Targeting of a heterologous protein to the NPC depended on determinants specifying SUMO-1 modification and also on additional determinants in the COOH-terminal domain of RanGAP1. SUMO-1 modification and these additional determinants were found to specify interaction between the COOH-terminal domain of RanGAP1 and a region of the nucleoporin, Nup358, between Ran-binding domains three and four. Together, these findings indicate that SUMO-1 modification targets RanGAP1 to the NPC by exposing, or creating, a Nup358 binding site in the COOH-terminal domain of RanGAP1. Surprisingly, the COOH-terminal domain of RanGAP1 was also found to harbor a nuclear localization signal. This nuclear localization signal, and the presence of nine leucine-rich nuclear export signal motifs, suggests that RanGAP1 may shuttle between the nucleus and the cytoplasm.
Figures
Figure 1
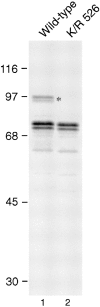
Lysine 526 is essential for SUMO-1 modification of RanGAP1 (A) Myc-tagged wild-type RanGAP1 (lane 1) and myc-tagged RanGAP1 containing a single lysine to arginine substitution at amino acid 526 (lane 2) were transcribed and translated in vitro in the presence of [35S]methionine and analyzed by SDS-PAGE. Molecular mass standards are indicated on the left and an asterisk denotes SUMO-1–modified wild-type RanGAP1.
Figure 3
Determinants for SUMO-1 modification reside in the COOH-terminal domain of RanGAP1. (A) Schematic representations of RanGAP1 and pyruvate kinase fusion proteins. Asterisks indicate the presence of a myc epitope tag at the NH2 terminus of each protein. Leucine-rich repeats in RanGAP1 are indicated by dark-shaded boxes, the acidic domain by a hatched box, and the COOH-terminal domain by a light-shaded box. Pyruvate kinase is represented by very light-shaded boxes, and SUMO-1 by a black box. Results of in vitro SUMO-1 modification and immunolocalization of the transiently expressed proteins are indicated on the right. C, cytoplasm; NE, nuclear envelope; N, nucleus. (B) Wild-type RanGAP1 (lane 1), the COOH-terminal deletion mutant CD23 (lane 2), the pyruvate kinase fusion proteins NΔ419/PK (lane 3), NΔ470/PK (lane 4) and NΔ502/PK (lane 5), and pyruvate kinase (lane 6) were transcribed and translated in rabbit reticulocyte extracts in the presence of [35S]methionine. Reactions were separated by SDS-PAGE and analyzed by autoradiography. Molecular mass standards are indicated on the left and asterisks indicate SUMO-1–modified substrates.
Figure 3
Determinants for SUMO-1 modification reside in the COOH-terminal domain of RanGAP1. (A) Schematic representations of RanGAP1 and pyruvate kinase fusion proteins. Asterisks indicate the presence of a myc epitope tag at the NH2 terminus of each protein. Leucine-rich repeats in RanGAP1 are indicated by dark-shaded boxes, the acidic domain by a hatched box, and the COOH-terminal domain by a light-shaded box. Pyruvate kinase is represented by very light-shaded boxes, and SUMO-1 by a black box. Results of in vitro SUMO-1 modification and immunolocalization of the transiently expressed proteins are indicated on the right. C, cytoplasm; NE, nuclear envelope; N, nucleus. (B) Wild-type RanGAP1 (lane 1), the COOH-terminal deletion mutant CD23 (lane 2), the pyruvate kinase fusion proteins NΔ419/PK (lane 3), NΔ470/PK (lane 4) and NΔ502/PK (lane 5), and pyruvate kinase (lane 6) were transcribed and translated in rabbit reticulocyte extracts in the presence of [35S]methionine. Reactions were separated by SDS-PAGE and analyzed by autoradiography. Molecular mass standards are indicated on the left and asterisks indicate SUMO-1–modified substrates.
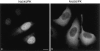
Figure 2
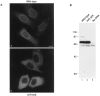
SUMO-1 modification is required for localization of RanGAP1 at the NPC. (A) HeLa cells were transfected with plasmids expressing myc-tagged wild-type RanGAP1 (a) or myc-tagged RanGAP1 containing a single lysine to arginine substitution at amino acid 526 (b). Subcellular localizations were determined 36 h after transfection by indirect immunofluorescence using the anti-myc monoclonal antibody, 9E10. (B) Cells were lysed in SDS sample buffer 36 h after transfection and analyzed by immunoblot analysis with the anti-myc monoclonal antibody, 9E10. Bar, 10 μm.
Figure 4
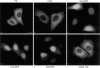
Pyruvate kinase is targeted to the NPC by SUMO-1 modification and by additional determinants in the COOH-terminal domain of RanGAP1. Plasmids coding for the proteins illustrated in Fig. 3_A_ were transfected into HeLa cells, and the transiently expressed proteins we immunolocalized with the anti-myc monoclonal antibody, 9E10. Representative micrographs of the observed immunolocalizations are presented. Bar, 10 μm.
Figure 5
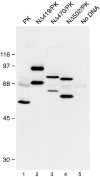
RanGAP1/PK fusion proteins are SUMO-1 modified in vivo. Cells transfected as in Fig. 4 were lysed in SDS sample buffer and analyzed by immunoblot analysis with the anti-myc monoclonal antibody, 9E10. In cells transfected with native pyruvate kinase, a single specific band migrating at 50 kD is detected (lane 1). The band migrating at 65 kD is nonspecific and detected in cells transfected with no DNA (lane 5). Two forms of the fusion proteins NΔ419/ PK (lane 2), NΔ470/PK (lane 3), and NΔ502/PK (lane 4) are detected, the lower molecular mass forms corresponding to unmodified protein, the higher molecular mass forms corresponding to SUMO-1–modified protein. Molecular mass standards are indicated on the left.
Figure 6
The COOH-terminal domain of RanGAP1 contains a nuclear localization signal. The pyruvate kinase fusion proteins NΔ540/PK (a) and NΔ555/PK (b) were transiently expressed in HeLa cells and localized with the anti-myc monoclonal antibody, 9E10.
Figure 7
RanGAP1 contains nine potential nuclear export signals in addition to a nuclear localization signal. (A) Schematic representation of RanGAP1 indicating the positions of the nine NH2-terminal nuclear export motifs and the COOH-terminal nuclear localization signal. (B) Amino acid sequence of the nuclear localization signal in the COOH-terminal domain of RanGAP1. (C) Amino acid sequences and alignments of the nine putative nuclear export motifs in the NH2-terminal domain of RanGAP1 and the nuclear export motifs of Rev (Fischer et al., 1995), PKI (Wen et al., 1995), IκB, and Rex (Fritz and Green, 1996). Hydrophobic residues comprising the core of the motif are highlighted in dark gray, and hydrophobic residues outside of the core are highlighted in light gray.
Figure 8
SUMO-1–modified RanGAP1 binds to a COOH-terminal region of Nup358, between Ran-binding domains three and four. (A) Schematic representation of Nup358. RBD, Ran-binding domain; CycH, cyclophilin homologous domain. Long vertical lines indicate FXFG repeats, and short vertical lines indicate FG repeats. Fragments that were found to bind modified RanGAP1 are indicated, as well as a summary of their binding activity (+, weak binding; ++, strong binding). Internal direct repeats in the RanGAP1-binding domain are indicated by open boxes. (B) Full-length RanGAP1 (lane 1) and COOH-terminal regions of RanGAP1 from amino acids 420–589 (NΔ419; lane 6) and 471– 589 (NΔ470; lane 11) were translated in vitro in the presence of [35S]methionine. Translated proteins were assayed for binding to the domains of Nup358 indicated in A, as described in Materials and Methods. Molecular mass standards are indicated on the left, and asterisks denote SUMO-1–modified proteins.
Figure 8
SUMO-1–modified RanGAP1 binds to a COOH-terminal region of Nup358, between Ran-binding domains three and four. (A) Schematic representation of Nup358. RBD, Ran-binding domain; CycH, cyclophilin homologous domain. Long vertical lines indicate FXFG repeats, and short vertical lines indicate FG repeats. Fragments that were found to bind modified RanGAP1 are indicated, as well as a summary of their binding activity (+, weak binding; ++, strong binding). Internal direct repeats in the RanGAP1-binding domain are indicated by open boxes. (B) Full-length RanGAP1 (lane 1) and COOH-terminal regions of RanGAP1 from amino acids 420–589 (NΔ419; lane 6) and 471– 589 (NΔ470; lane 11) were translated in vitro in the presence of [35S]methionine. Translated proteins were assayed for binding to the domains of Nup358 indicated in A, as described in Materials and Methods. Molecular mass standards are indicated on the left, and asterisks denote SUMO-1–modified proteins.
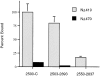
Figure 9
NΔ419 and NΔ470 have different affinities for the COOH-terminal region of Nup358 between Ran-binding domains three and four. Binding assays and quantification were performed as described in Material and Methods. Mean values are shown, with error bars indicating variance observed between triplicate experiments.
Similar articles
- A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2.
Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. Mahajan R, et al. Cell. 1997 Jan 10;88(1):97-107. doi: 10.1016/s0092-8674(00)81862-0. Cell. 1997. PMID: 9019411 - A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex.
Matunis MJ, Coutavas E, Blobel G. Matunis MJ, et al. J Cell Biol. 1996 Dec;135(6 Pt 1):1457-70. doi: 10.1083/jcb.135.6.1457. J Cell Biol. 1996. PMID: 8978815 Free PMC article. - Ubc9p and the conjugation of SUMO-1 to RanGAP1 and RanBP2.
Saitoh H, Sparrow DB, Shiomi T, Pu RT, Nishimoto T, Mohun TJ, Dasso M. Saitoh H, et al. Curr Biol. 1998 Jan 15;8(2):121-4. doi: 10.1016/s0960-9822(98)70044-2. Curr Biol. 1998. PMID: 9427648 - Nuclear import and export pathways.
Moroianu J. Moroianu J. J Cell Biochem. 1999;Suppl 32-33:76-83. doi: 10.1002/(sici)1097-4644(1999)75:32+<76::aid-jcb10>3.3.co;2-h. J Cell Biochem. 1999. PMID: 10629106 Review. - Nucleocytoplasmic transport: driving and directing transport.
Cole CN, Hammell CM. Cole CN, et al. Curr Biol. 1998 May 21;8(11):R368-72. doi: 10.1016/s0960-9822(98)70239-8. Curr Biol. 1998. PMID: 9635180 Review.
Cited by
- Poly-SUMO-2/3 chain modification of Nuf2 facilitates CENP-E kinetochore localization and chromosome congression during mitosis.
Subramonian D, Chen TA, Paolini N, Zhang XD. Subramonian D, et al. Cell Cycle. 2021 May;20(9):855-873. doi: 10.1080/15384101.2021.1907509. Epub 2021 Apr 28. Cell Cycle. 2021. PMID: 33910471 Free PMC article. - An acetylation switch regulates SUMO-dependent protein interaction networks.
Ullmann R, Chien CD, Avantaggiati ML, Muller S. Ullmann R, et al. Mol Cell. 2012 Jun 29;46(6):759-70. doi: 10.1016/j.molcel.2012.04.006. Epub 2012 May 10. Mol Cell. 2012. PMID: 22578841 Free PMC article. - Evolution of a signalling system that incorporates both redundancy and diversity: Arabidopsis SUMOylation.
Chosed R, Mukherjee S, Lois LM, Orth K. Chosed R, et al. Biochem J. 2006 Sep 15;398(3):521-9. doi: 10.1042/BJ20060426. Biochem J. 2006. PMID: 16740136 Free PMC article. - The human Nup107-160 nuclear pore subcomplex contributes to proper kinetochore functions.
Zuccolo M, Alves A, Galy V, Bolhy S, Formstecher E, Racine V, Sibarita JB, Fukagawa T, Shiekhattar R, Yen T, Doye V. Zuccolo M, et al. EMBO J. 2007 Apr 4;26(7):1853-64. doi: 10.1038/sj.emboj.7601642. Epub 2007 Mar 15. EMBO J. 2007. PMID: 17363900 Free PMC article. - Biology and biophysics of the nuclear pore complex and its components.
Lim RY, Ullman KS, Fahrenkrog B. Lim RY, et al. Int Rev Cell Mol Biol. 2008;267:299-342. doi: 10.1016/S1937-6448(08)00632-1. Int Rev Cell Mol Biol. 2008. PMID: 18544502 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous