The yersiniabactin biosynthetic gene cluster of Yersinia enterocolitica: organization and siderophore-dependent regulation - PubMed (original) (raw)
The yersiniabactin biosynthetic gene cluster of Yersinia enterocolitica: organization and siderophore-dependent regulation
C Pelludat et al. J Bacteriol. 1998 Feb.
Abstract
The ability to synthesize and uptake the Yersinia siderophore yersiniabactin is a hallmark of the highly pathogenic, mouse-lethal species Yersinia pestis, Y. pseudotuberculosis, and Y. enterocolitica 1B. We have identified four genes, irp1, irp3, irp4, and irp5, on a 13-kb chromosomal DNA fragment of Y. enterocolitica 08, WA-314. These genes constitute the yersiniabactin biosynthetic gene cluster together with the previously defined irp2. The irp1 gene consists of 9,486 bp capable of encoding a 3,161-amino-acid high-molecular-weight protein 1 (HMWP1) polypeptide with a predicted mass of 384.6 kDa. The first 3,000 bp of irp1 show similarity to the corresponding regions of the polyketide synthase genes of Bacillus subtilis and Streptomyces antibioticus. The remaining part of irp1 is most similar to irp2, encoding HMWP2, which might be the reason for immunological cross-reactivity of the two polypeptides. Irp4 was found to have 41.7% similarity to thioesterase-like protein of the anguibactin biosynthetic genes of Vibrio anguillarum. Irp5 shows 41% similarity to EntE, the 2,3-dihydroxybenzoic acid-activating enzyme utilized in enterobactin synthesis of Escherichia coli. Irp4 and Irp5 are nearly identical to YbtT and YbtE, recently identified in Y. pestis. irp3 has no similarity to any known gene. Inactivation of either irp1 or irp2 abrogates yersiniabactin synthesis. Mutations in irp1 or fyuA (encoding yersiniabactin/pesticin receptor) result in downregulation of irp2 that can be upregulated by the addition of yersiniabactin. A FyuA-green fluorescent protein translational fusion was downregulated in an irp1 mutant. Upregulation was achieved by addition of yersiniabactin but not desferal, pesticin, or pyochelin, which indicates high specificity of the FyuA receptor and autoregulation of genes involved in synthesis and uptake of yersiniabactin.
Figures
FIG. 1
Genetic organization of the Y. enterocolitica O8 WA-314 irp2-fyuA gene cluster. The genes are depicted as boxes. Arrows above indicate the direction of transcription. E, _Eco_RI; S, _Sal_I.
FIG. 2
Common amino acids motifs found in HMWP1 and HMWP2.
FIG. 3
Southern hybridization of chromosomal DNAs from Yersinia and E. coli strains with the irp1 probe. The chromosomal DNA was digested with Eco_RI, and the resulting fragments were separated on a 1% agarose gel prior to Southern blotting. Hybridization was performed with a DIG-labeled PCR probe generated with primers i8513 and i8730. Lane 1, Y. pestis KUMA; lane 2, Y. pestis KIM; lane 3, Y. pestis KIM Δ_pgm; lane 4, Y. pseudotuberculosis 346; lane 5, Y. pseudotuberculosis 201; lane 6, Y. pseudotuberculosis PB1; lane 7, Y. enterocolitica 8081; lane 8, Y. enterocolitica WA-CS; lane 9, Y. enterocolitica Y5.27; lane 10, Y. enterocolitica Y-96-C; lane 11, Y. enterocolitica Y-108-C; lane 12, E. coli Phi; lane 13; E. coli DH5α.
FIG. 4
Comparison of the region from bp 1041 to 1191 of irp1 in Y. enterocolitica 8081, Y. enterocolitica WA-CS (I), Y. pestis KUMA, Y. pestis KIM, Y. pseudotuberculosis 346, Y. pseudotuberculosis PB1, and E. coli Phi (II). Nonmatching bases are boldfaced and underlined.
FIG. 5
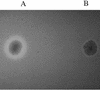
CAS agar plate showing siderophore-producing Y. enterocolitica WA-CS (A; with halo) and mutant WA-CS irp1::Kanr (B).
FIG. 6
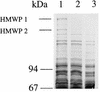
Expression of HMWP1 and -2 in WA-CS irp1::Kanr and WA fyuA mutants. SDS-PAGE (7.5% gel) of total-cell proteins from iron-starved strains WA-CS (lane 1), WA-CS irp1::Kanr (lane 2), and WA fyuA (lane 3).
FIG. 7
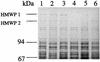
SDS-PAGE (7.5% gel) (A) and corresponding immunoblot (B) with cell lysates of WA-CS (lane 1), WA-CS irp1::Kanr (lane 2), and WA-CS irp1::Kanr with an addition of purified yersiniabactin (lane 3). The strains were grown under iron starvation in NBD medium. Western blotting was performed with HMWP polyclonal antibodies kindly provided from Elisabeth Carniel.
FIG. 8
Effects of pesticin and yersiniabactin on expression of the HMWPs. SDS-PAGE (7.5% gel) of total-cell proteins of iron-starved strains WA-CS and WA-CS irp1::Kanr with addition of pesticin and culture supernatant containing yersiniabactin. Lane 1, WA-CS in NBD medium; lane 2, WA-CS in NBD medium with pesticin (1:1,024); lane 3, WA-CS in NBD medium with yersiniabactin supernatant (1:50); lane 4, WA-CS irp1::Kanr in NBD medium; lane 5, WA-CS irp1::Kanr in NBD medium with pesticin; lane 6, WA-CS irp1::Kanr in NBD medium with yersiniabactin supernatant.
FIG. 9
Fluorescence of FyuA-GFP in Y. enterocolitica WA-CS (- -) and WA-CS irp1::Kanr (—) in NB medium (A), NBD medium (B), NBD medium plus purified yersiniabactin (C), and NBD medium plus desferrioxamine B (D). (E) Positive (WA-CS[pGFP mut 3]; -- --) and negative (WA-CS; -- —— --) controls. Au, arbitrary units.
FIG. 10
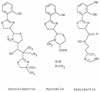
Chemical structures of yersiniabactin, pyochelin, and anguibactin.
Similar articles
- Hunger for iron: the alternative siderophore iron scavenging systems in highly virulent Yersinia.
Rakin A, Schneider L, Podladchikova O. Rakin A, et al. Front Cell Infect Microbiol. 2012 Nov 30;2:151. doi: 10.3389/fcimb.2012.00151. eCollection 2012. Front Cell Infect Microbiol. 2012. PMID: 23226687 Free PMC article. Review. - Functional analysis of yersiniabactin transport genes of Yersinia enterocolitica.
Brem D, Pelludat C, Rakin A, Jacobi CA, Heesemann J. Brem D, et al. Microbiology (Reading). 2001 May;147(Pt 5):1115-1127. doi: 10.1099/00221287-147-5-1115. Microbiology (Reading). 2001. PMID: 11320115 - The pesticin receptor of Yersinia enterocolitica: a novel virulence factor with dual function.
Rakin A, Saken E, Harmsen D, Heesemann J. Rakin A, et al. Mol Microbiol. 1994 Jul;13(2):253-63. doi: 10.1111/j.1365-2958.1994.tb00420.x. Mol Microbiol. 1994. PMID: 7984105 - Evidence for two evolutionary lineages of highly pathogenic Yersinia species.
Rakin A, Urbitsch P, Heesemann J. Rakin A, et al. J Bacteriol. 1995 May;177(9):2292-8. doi: 10.1128/jb.177.9.2292-2298.1995. J Bacteriol. 1995. PMID: 7730256 Free PMC article. - The Yersinia high-pathogenicity island: an iron-uptake island.
Carniel E. Carniel E. Microbes Infect. 2001 Jun;3(7):561-9. doi: 10.1016/s1286-4579(01)01412-5. Microbes Infect. 2001. PMID: 11418330 Review.
Cited by
- Carbapenemase-Producing Klebsiella pneumoniae From Transplanted Patients in Brazil: Phylogeny, Resistome, Virulome and Mobile Genetic Elements Harboring bla KPC- 2 or bla NDM- 1.
Raro OHF, da Silva RMC, Filho EMR, Sukiennik TCT, Stadnik C, Dias CAG, Oteo Iglesias J, Pérez-Vázquez M. Raro OHF, et al. Front Microbiol. 2020 Jul 15;11:1563. doi: 10.3389/fmicb.2020.01563. eCollection 2020. Front Microbiol. 2020. PMID: 32760368 Free PMC article. - Hunger for iron: the alternative siderophore iron scavenging systems in highly virulent Yersinia.
Rakin A, Schneider L, Podladchikova O. Rakin A, et al. Front Cell Infect Microbiol. 2012 Nov 30;2:151. doi: 10.3389/fcimb.2012.00151. eCollection 2012. Front Cell Infect Microbiol. 2012. PMID: 23226687 Free PMC article. Review. - The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000.
Buell CR, Joardar V, Lindeberg M, Selengut J, Paulsen IT, Gwinn ML, Dodson RJ, Deboy RT, Durkin AS, Kolonay JF, Madupu R, Daugherty S, Brinkac L, Beanan MJ, Haft DH, Nelson WC, Davidsen T, Zafar N, Zhou L, Liu J, Yuan Q, Khouri H, Fedorova N, Tran B, Russell D, Berry K, Utterback T, Van Aken SE, Feldblyum TV, D'Ascenzo M, Deng WL, Ramos AR, Alfano JR, Cartinhour S, Chatterjee AK, Delaney TP, Lazarowitz SG, Martin GB, Schneider DJ, Tang X, Bender CL, White O, Fraser CM, Collmer A. Buell CR, et al. Proc Natl Acad Sci U S A. 2003 Sep 2;100(18):10181-6. doi: 10.1073/pnas.1731982100. Epub 2003 Aug 19. Proc Natl Acad Sci U S A. 2003. PMID: 12928499 Free PMC article. - Rational live oral carrier vaccine design by mutating virulence-associated genes of Yersinia enterocolitica.
Igwe EI, Rüssmann H, Roggenkamp A, Noll A, Autenrieth IB, Heesemann J. Igwe EI, et al. Infect Immun. 1999 Oct;67(10):5500-7. doi: 10.1128/IAI.67.10.5500-5507.1999. Infect Immun. 1999. PMID: 10496939 Free PMC article. - Possible use of ail and foxA polymorphisms for detecting pathogenic Yersinia enterocolitica.
Huang Y, Wang X, Cui Z, Yang Y, Xiao Y, Tang L, Kan B, Xu J, Jing H. Huang Y, et al. BMC Microbiol. 2010 Aug 7;10:211. doi: 10.1186/1471-2180-10-211. BMC Microbiol. 2010. PMID: 20691098 Free PMC article.
References
- Aparicio J F, Molnar I, Schwecke T, Konig A, Haydock S F, Khaw L E, Staunton J, Leadlay P F. Organization of the biosynthetic gene cluster for rapamycin in Streptomyces hygroscopicus: analysis of the enzymatic domains in the modular polyketide synthase. Gene. 1996;169:9–16. - PubMed
- Baumler A, Koebnik R, Stojiljkovic I, Heesemann J, Braun V, Hantke K. Survey of newly characterized iron uptake systems of Yersinia enterocolitica. Int J Med Microbiol Virol Parasitol Infect Dis. 1993;278:416–424. - PubMed
- Ben Gurion R, Shafferman A. Essential virulence determinants of different Yersinia species are carried on a common plasmid. Plasmid. 1981;5:183–187. - PubMed
- Berkovier H, Mollaret H H. The family Enterobacteriaceae, genus Yersinia. In: Krieg N R, Holt J H, editors. Bergey’s manual of systematic bacteriology. Baltimore, Md: The Williams & Wilkins Co.; 1984. pp. 498–506.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials