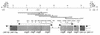Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation - PubMed (original) (raw)
Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation
U Römling et al. J Bacteriol. 1998 Feb.
Abstract
Mouse-virulent Salmonella typhimurium strains SR-11 and ATCC 14028-1s express curli fibers, thin aggregative fibers, at ambient temperature on plates as judged by Western blot analysis and electron microscopy. Concomitantly with curli expression, cells develop a rough and dry colony morphology and bind the dye Congo red (called the rdar morphotype). Cloning and characterization of the two divergently transcribed operons required for curli biogenesis, csgBA(C) and csgDEFG, from S. typhimurium SR-11 revealed the same gene order and flanking genes as in Escherichia coli. The divergence of the curli region between S. typhimurium and E. coli at the nucleotide level is above average (22.4%). However, a high level of conservation at the protein level, which ranged from 86% amino acid homology for the fiber subunit CsgA to 99% homology for the lipoprotein CsgG, implies functional constraints on the gene products. Consequently, S. typhimurium genes on low-copy-number plasmids were able to complement respective E. coli mutants, although not always to wild-type levels. rpoS and ompR are required for transcriptional activation of (at least) the csgD promoter. The high degree of conservation at the protein level and the identical regulation patterns in E. coli and S. typhimurium suggest similar roles of curli fibers in the same ecological niche in the two species.
Figures
FIG. 1
Colony morphology and color of S. typhimurium ATCC 14028-1s and SR-11 and their respective derivatives. (A) Cells grown at 28°C on LB medium plates without salt containing the dye CR. SR-11 and ATCC 14028-1s (here shown as the Nalr derivatives UMR3 and UMR1, respectively) developed a rough colony morphology with a dry surface and showed a deep red color by binding the dye CR, the rdar morphotype. The strains with deletions in csgA (MAE1, MAE2, and MAE5) were also rough but had a shinier surface. Binding of the dye CR led to a pinkish color of the colonies (the pdar morphotype). The strains with deletions in rpoS (MAE40 and MAE29) and ompR (MAE46 and MAE34) were white. (B) The same strains as in panel A but grown at 37°C. All cells were white and smooth, the saw morphotype.
FIG. 2
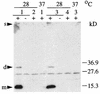
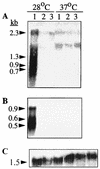
Western blot analysis of fiber-derived CsgA from whole cells grown at 28 and 37°C. The cell pellets were immediately resuspended in SDS sample buffer (−) or treated with formic acid (+) as described in Materials and Methods. Minor signals for the fiber subunit which could vary in their intensities were regularly found in the slot (s) and in the gel corresponding to a dimer (d), while the major signal was consistent with the running behavior of a monomer (m). SR-11 and ATCC 14024-1s showed a signal for CsgA only at 28°C and not at 37°C. csgA knockouts did not show any signal at all. Lanes: 1, SR-11; 2, MAE2; 3, ATCC 14028-1s; 4, MAE1.
FIG. 3
Electron micrograph of negatively stained ATCC 14028-1s cells grown at 28°C on LB agar without salt. The cells which are surrounded by a thick layer of curli fibers show a different cell morphology. Bar, 1 μm.
FIG. 4
Organization of the csg region on the S. typhimurium SR-11 chromosome. A restriction map containing sites important for the cloning and localization of the csg region is shown. The positions of the genes required for curli biogenesis, csgDEFG and csgBAC (boxes), the position of the transcriptional start sites and the direction of polymerization (flags), and the position of the promoter (circle) are indicated. Experimentally confirmed RNA full-length transcripts (arrows above gene boxes) and open reading frames for which transcriptional analysis had not been carried out or for which no transcript had been detected so far (boxes with arrowheads inside) are also shown. The DNA fragments used as probes in RNA transcript analysis (dotted lines), primers used for the PCR amplification of DNA fragments (arrowheads above dotted lines), and subclones used to complement E. coli isolates with mutations in the csg genes (pCSGA, pCSGB, pCSGD, pCSGE, pCSGF, pCSGG, and pCSGEFG) (bars with small arrows indicating the transcription from the lacZ promoter) are indicated. B, _Bst_YI; E, _Eco_RI; H, _Hin_dIII; N, _Nsi_I; P, _Pst_I; S, _Sac_I.
FIG. 5
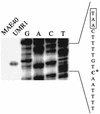
Primer extension analyses for the determination of the transcriptional start site of the csgDEFG operon. RNA was prepared from strains UMR1 and MAE40 (the rpoS derivative of ATCC 14028-1s) grown at 28°C on plates, and primer extension was carried out as described in Materials and Methods. An extension product is seen for UMR1 but not for MAE40. Primer PEXD1, located 58 bp downstream of the csgDEFG start codon, was used for the extension reaction as well as for the sequencing reaction on pUMR10-7 as a template. The sequence derived from the PEXD1 primer on pUMR10-7 is complementary to the RNA template, so the coding strand is automatically shown. The transcriptional start site (asterisk) and bases belonging to the putative promoter sequences (boxed) are indicated.
FIG. 6
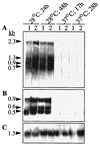
Northern blot analysis of RNA transcripts of the csg region using S. typhimurium ATCC 14028-1s and SR-11 grown on LB medium plates without salt at 28 and 37°C for different periods. Probes covering the whole csgD and csgA genes were used; their locations are indicated in Fig. 4. Lanes: 1, UMR3; 2, UMR1. The sizes of the bands detected by the respective probes are shown on the left, calculated by using a 0.24- to 9.5-kb RNA ladder (Life Technologies) as a standard. (A) Hybridization with the csgD probe; (B) hybridization with the csgA probe; (C) control hybridization with 16S RNA as described in Materials and Methods.
FIG. 7
Western blot analysis of the complementation of the E. coli csg mutants. Results for whole-cell preparations treated with formic acid are shown. The mutant (−) and the mutant harboring the respective complementing plasmid (+) were run next to each other. Except for MHR480 (Δ_csgE3_), none of the E. coli mutants showed a CsgA signal derived from polymerized fibers. MHR261 (csgB2) and MHR503 (csgD6) were complemented to wild-type levels by using plasmids with the single genes. MHR426 (csgF4) was complemented to wild type only when a plasmid carrying csgEFG was used. Faint signals of CsgA were detected when pCSGA was introduced into MHR204 (csgA::Tn_105_).
FIG. 8
Northern blot analysis of RNA transcripts from S. typhimurium ATCC 14028-1s and rpoS and ompR derivatives grown on LB medium plates without salt at 28 and 37°C. Probes covering the whole csgD and csgA genes were used; their locations are indicated in Fig. 4. Lanes: 1, UMR1; 2, MAE40 (rpoS); 3, MAE46 (ompR). The sizes of the bands detected by the respective probes are shown on the left, calculated by using a 0.24- to 9.5-kb RNA ladder (Life Technologies) as a standard. (A) Hybridization with the csgD probe; (B) hybridization with the csgA probe; (C) control hybridization with 16S RNA as described in Materials and Methods.
Similar articles
- MlrA, a novel regulator of curli (AgF) and extracellular matrix synthesis by Escherichia coli and Salmonella enterica serovar Typhimurium.
Brown PK, Dozois CM, Nickerson CA, Zuppardo A, Terlonge J, Curtiss R 3rd. Brown PK, et al. Mol Microbiol. 2001 Jul;41(2):349-63. doi: 10.1046/j.1365-2958.2001.02529.x. Mol Microbiol. 2001. PMID: 11489123 - Expression of two csg operons is required for production of fibronectin- and congo red-binding curli polymers in Escherichia coli K-12.
Hammar M, Arnqvist A, Bian Z, Olsén A, Normark S. Hammar M, et al. Mol Microbiol. 1995 Nov;18(4):661-70. doi: 10.1111/j.1365-2958.1995.mmi_18040661.x.. Mol Microbiol. 1995. PMID: 8817489 - Multicellular and aggregative behaviour of Salmonella typhimurium strains is controlled by mutations in the agfD promoter.
Römling U, Sierralta WD, Eriksson K, Normark S. Römling U, et al. Mol Microbiol. 1998 Apr;28(2):249-64. doi: 10.1046/j.1365-2958.1998.00791.x. Mol Microbiol. 1998. PMID: 9622351 - Curli provide the template for understanding controlled amyloid propagation.
Wang X, Chapman MR. Wang X, et al. Prion. 2008 Apr-Jun;2(2):57-60. doi: 10.4161/pri.2.2.6746. Epub 2008 Apr 5. Prion. 2008. PMID: 19098444 Free PMC article. Review.
Cited by
- Amyloids: The History of Toxicity and Functionality.
Yakupova EI, Bobyleva LG, Shumeyko SA, Vikhlyantsev IM, Bobylev AG. Yakupova EI, et al. Biology (Basel). 2021 May 1;10(5):394. doi: 10.3390/biology10050394. Biology (Basel). 2021. PMID: 34062910 Free PMC article. Review. - Identification of novel genes involved in the biofilm formation process of Avian Pathogenic Escherichia coli (APEC).
Young MM, de Oliveira AL, Nolan LK, Barbieri NL, Logue CM. Young MM, et al. PLoS One. 2022 Dec 19;17(12):e0279206. doi: 10.1371/journal.pone.0279206. eCollection 2022. PLoS One. 2022. PMID: 36534660 Free PMC article. - The Escherichia coli tppB (ydgR) gene represents a new class of OmpR-regulated genes.
Goh EB, Siino DF, Igo MM. Goh EB, et al. J Bacteriol. 2004 Jun;186(12):4019-24. doi: 10.1128/JB.186.12.4019-4024.2004. J Bacteriol. 2004. PMID: 15175316 Free PMC article. - Genetic diversity and virulence characteristics of biofilm-producing uropathogenic Escherichia coli.
Qasemi A, Rahimi F, Katouli M. Qasemi A, et al. Int Microbiol. 2022 May;25(2):297-307. doi: 10.1007/s10123-021-00221-w. Epub 2021 Oct 27. Int Microbiol. 2022. PMID: 34705131 - Linkage map of Escherichia coli K-12, edition 10: the traditional map.
Berlyn MK. Berlyn MK. Microbiol Mol Biol Rev. 1998 Sep;62(3):814-984. doi: 10.1128/MMBR.62.3.814-984.1998. Microbiol Mol Biol Rev. 1998. PMID: 9729611 Free PMC article. Review.
References
- Applied Biosystems, Inc. User bulletin 18. Foster City, Calif: Applied Biosystems, Inc.; 1991.
- Arnqvist A, Olsén A, Normark S. ςS-dependent growth-phase induction of the csgBA promoter can be achieved in vivo by ς70 in the absence of the nucleoid-associated protein H-NS. Mol Microbiol. 1994;13:1021–1032. - PubMed
- Arnqvist A, Olsén A, Pfeifer J, Russell D G, Normark S. The Crl protein activates cryptic genes for curli formation and fibronectin binding in Escherichia coli HB101. Mol Microbiol. 1992;6:2443–2452. - PubMed
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1994.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials