Reverse transcriptase (RT) inhibition of PCR at low concentrations of template and its implications for quantitative RT-PCR - PubMed (original) (raw)
Reverse transcriptase (RT) inhibition of PCR at low concentrations of template and its implications for quantitative RT-PCR
D P Chandler et al. Appl Environ Microbiol. 1998 Feb.
Abstract
Numerous instances of reverse transcriptase (RT) inhibition of the PCR were observed while developing nonquantitative uncoupled RT-PCR techniques for detecting nitrogenase and ammonia monooxygenase gene expression in situ. The inhibitory effect of RT on the PCR was removed with increasing template concentrations beyond 10(5) to 10(6) copies. Including T4 gene 32 protein during the reverse transcription phase of the RT-PCR reaction increased the RT-PCR product yield by as much as 483%; if gene 32 protein was introduced after reverse transcription but prior to the PCR phase, no improvement in product yield was observed. Addition of 1 microgram of exogenous calf thymus DNA or yeast tRNA did little to relieve RT inhibition of the PCR on both genomic DNA and mRNA templates. These results suggest that RT inhibition of the PCR is mediated through direct interaction with the specific primer-template combination (DNA and RNA) and point to specific assay modifications for estimating the extent of RT inhibition and counteracting some of the inhibitory effect. Furthermore, the working hypothesis of RT inhibition below a 10(5) to 10(6) copy threshold has important implications for quantitative RT-PCR studies. In particular, competitive, quantitative RT-PCR systems will consistently underestimate the actual RNA concentration. Hence, enumerations of RNA templates below 10(5) to 10(6) copies will be relative to an internal standard and will not be an absolute measure of RNA abundance in situ.
Figures
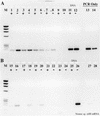
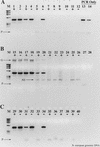
FIG. 1
RT inhibition of the PCR. +, RT present; −, RT absent. Lanes: M, φX174 × _Hae_III molecular weight marker; 1 and 2, no template; 3 and 4, 200 fg of N. europaea genomic DNA; 5 and 6, 2 pg; 7 and 8, 20 pg; 9 and 10, 20 pg; 11 and 12, 2 ng; 13, 200 fg of N. europaea genomic DNA amplified by PCR only; 14, 2 pg of N. europaea genomic DNA; 15, 20 pg of N. europaea genomic DNA.
FIG. 2
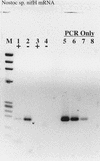
RT inhibition of RT-PCR demonstrated on Nostoc sp. nifH mRNA. Lanes: M, φX174 × _Hae_III molecular weight marker; 1, 5 μl of purified mRNA plus RT; 2, 5 μl of purified mRNA without RT; 3 and 4, same as 1 and 2, except that the mRNA sample was pretreated with RNase A prior to RT-PCR; 5, 20 pg of Nostoc sp. genomic DNA subject to PCR amplification only; 6, 2 pg of genomic DNA; 7, 200 fg of genomic DNA; 8, no-template control.
FIG. 3
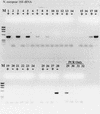
RT-PCR with a dilution series of purified N. europaea 16S rRNA as template. +, RT present; −, RT absent. Lanes: M, φX174 × _Hae_III molecular weight marker; 1 and 2, 2 ng of 16S rRNA with or without RT; 3 and 4, 200 pg; 5 and 6, 20 pg; 7 and 8, 2 pg; 9 and 10, 200 fg; 11 and 12, 20 fg; 13 and 14, 2 fg; 15 and 16, no-template control; 17 and 18, 2 ng of N. europaea genomic DNA; 19 to 24, same as lanes 1 to 6, except that the sample was pretreated with RNase A prior to RT-PCR; 25 and 26, no-template controls pretreated with RNase A; 27 and 28, 2 ng of N. europaea genomic DNA pretreated with RNase A; 29, 20 pg of genomic DNA, PCR-only control; 30, 2 pg of genomic DNA; 31, 200 fg of genomic DNA; 32, no-template PCR-only control.
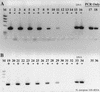
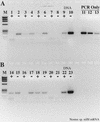
FIG. 4
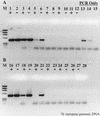
Relief of RT-PCR inhibition through the use of T4 gene 32 protein. N. europaea genomic DNA was used as template. (A) With T4 gene 32 protein; (B) without T4 gene 32 protein. +, RT present; −, RT absent. Lanes: M, φX174 × _Hae_III molecular weight marker; 1 and 2, 2 ng of DNA with or without RT; 3 and 4, 200 pg; 5 and 6, 20 pg; 7 and 8, 2 pg; 9 and 10, 200 fg; 11 and 12, no-template RT-PCR controls; 13 to 15 = 20, 2, and 0.2 pg of genomic DNA PCR controls; 16 to 27, same as lanes 1 to 12, except without gene 32 protein; 28, no-template PCR control.
FIG. 5
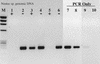
Relief of RT inhibition during the reverse transcription step in RT-PCR. Nostoc genomic DNA (200 pg) was used as the template. T4 gene 32 protein was added during the RT phase of RT-PCR or only for the PCR portion of the RT-PCR protocol. +, RT present; −, RT absent. Lanes: M, φX174 × _Hae_III molecular weight marker; 1 and 2, no added T4 gene 32 protein; 3 and 4, gene 32 protein added during the RT step; 5 and 6, gene 32 protein added for the PCR phase only; 7, 20 pg of genomic DNA, PCR only; 8, 2 pg of DNA; 9, 200 fg of DNA; 10, no-template PCR control.
FIG. 6
Effect of T4 gene 32 protein on RT-PCR amplification of Nostoc nifH mRNA. (A) Samples included 1.5 μg of gene 32 protein per RT-PCR; (B) no gene 32 protein present. +, RT present; −, RT absent. Lanes: M, φX174 × _Hae_III molecular weight marker; 1, 2, 15, and 16, 5 μl of mRNA as template; 3, 4, 17, and 18, 2 μl of mRNA; 5, 6, 19, and 20, 1 μl of mRNA; 7, 8, 21, and 22, 0.5 μl of mRNA; 9, 10, 23, and 24, RT-PCR no-template controls; 11, 12, 25, and 26, RT-PCR 200 pg of Nostoc genomic DNA controls; 13, 14, 27, and 28, PCR-only controls; 13, 20 pg of genomic DNA; 14, 2 pg; 27, 200 fg, 28, no template.
FIG. 7
Effect of T4 gene 32 protein on RT-PCR amplification of N. europaea 16S rRNA. (A) Samples included 1.5 μg of gene 32 protein per RT-PCR; (B) no gene 32 protein. +, RT present; −, RT absent. Lanes: M, φX174 × _Hae_III molecular weight marker; 1, 2, 19, and 20, 2 ng of 16S rRNA as template; 3, 4, 21, and 22, 200 pg; 5, 6, 23, and 24, 20 pg; 7, 8, 25, and 26, 2 pg; 9, 10, 27, and 28, 200 fg; 11, 12, 29, and 30, 20 fg; 13, 14, 31, and 32, RT-PCR no-template controls; 15, 16, 33, and 34, 200 pg of N. europaea genomic DNA RT-PCR controls; 17, 18, 35, and 36, PCR-only controls; 17, 20 pg of N. europaea genomic DNA; 18, 2 pg of DNA; 35, 200 fg of DNA; 36, PCR-only no-template control.
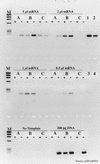
FIG. 8
Specificity of T4 gene 32 protein interaction during RT-PCR amplification of Nostoc nifH mRNA. +, RT present; −, RT absent. Lanes: M, φX174 × _Hae_III molecular weight marker. Lanes were assigned based upon template identification, as indicated above each block of samples. Samples were also paired according to T4 gene 32 protein treatment; A, no-gene 32 protein; B, gene 32 protein added during the reverse transcription step of RT-PCR; C, gene 32 protein added after the reverse transcription step, and beginning of PCR. PCR-only controls were Nostoc genomic DNA; lane 1, 20 pg; lane 2, 2 pg; lane 3, 200 fg; lane 4, no template.
FIG. 9
Effect of T4 gene 32 protein on RT-PCR amplification of Nostoc nifH mRNA isolated from cryptogamic crust. (A) Samples included 1.5 μg of gene 32 protein per RT-PCR; (B) no gene 32 protein. +, RT present; −, RT absent. Lanes: M, φX174 × _Hae_III molecular weight marker; 1, 2, 15, and 16, 10 μl of mRNA as template; 3, 4, 17, and 18, 5 μl of mRNA; 5, 6, 19, and 20, 2 μl of mRNA; 7, 8, 21, and 22, no-template RT-PCR controls; 9, 10, 23, and 24, 200 pg of Nostoc genomic DNA RT-PCR controls; 11 to 14, PCR-only controls; 11, 20 pg of Nostoc genomic DNA; 12, 2 pg; 13, 200 fg; 14, no template.
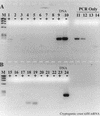
FIG. 10
Effects of nonspecific, noncompetitive templates on RT inhibition of the PCR. N. europaea genomic DNA was used as the template. (A) No added competitive template; (B) 1 μg of calf thymus DNA added during the reverse transcription step; (C) 1 μg of yeast tRNA added during the reverse transcription step. +, RT present; −, RT absent. Lanes: M, φX174 × _Hae_III molecular weight marker; 1 and 2, 2 ng of DNA; 3 and 4, 200 pg; 5 and 6, 20 pg; 7 and 8, 2 pg; 9 and 10, 200 fg; 11 and 12, no template; 13, 20 pg of DNA, PCR-only control; 14, 2 pg of DNA, PCR-only control; 15 to 26, same as lanes 1 to 12 except with calf thymus DNA as nonspecific template; 27, 200 fg of DNA PCR-only control; 28, no-template PCR-only control; 29 to 40, same as lanes 1 to 12 except with yeast tRNA as the nonspecific template. Arrows show the position of calf thymus genomic DNA (G), a nonspecific amplification product resulting from calf thymus genomic DNA (NS), and primer artifacts (P).
FIG. 11
Effects of nonspecific template on RT-PCR from Nostoc nifH mRNA. (A) No added nonspecific template; (B) 1 μg of yeast tRNA added during the reverse transcription step. +, RT present; −, RT absent. Lanes: M, φX174 × _Hae_III molecular weight marker; 1 and 2, 2 μl of nifH mRNA template; 3 and 4, 1 μl; 5 and 6, 0.5 μl; 7 and 8, no template; 9 and 10, 1 ng of Nostoc sp. genomic DNA, RT-PCR control; 11, 20 pg of genomic DNA, PCR-only control; 12, 2 pg of DNA; 13, 200 fg of DNA; 14, no-template PCR-only control; 15 to 23, same as lanes 1 to 10 except with yeast tRNA during the reverse transcription step.
Similar articles
- Increased yield of PCR products by addition of T4 gene 32 protein to the SMART PCR cDNA synthesis system.
Villalva C, Touriol C, Seurat P, Trempat P, Delsol G, Brousset P. Villalva C, et al. Biotechniques. 2001 Jul;31(1):81-3, 86. doi: 10.2144/01311st04. Biotechniques. 2001. PMID: 11464524 - [Use of the real-time RT-PCR method for investigation of small stable RNA expression level in human epidermoid carcinoma cells A431].
Nikitina TV, Nazarova NIu, Tishchenko LI, Tuohimaa P, Sedova VM. Nikitina TV, et al. Tsitologiia. 2003;45(4):392-402. Tsitologiia. 2003. PMID: 14520871 Russian. - Elimination of background signals in a modified polymerase chain reaction-based reverse transcriptase assay.
Maudru T, Peden K. Maudru T, et al. J Virol Methods. 1997 Jul;66(2):247-61. doi: 10.1016/s0166-0934(97)00067-0. J Virol Methods. 1997. PMID: 9255736 - Strand transfer events during HIV-1 reverse transcription.
Basu VP, Song M, Gao L, Rigby ST, Hanson MN, Bambara RA. Basu VP, et al. Virus Res. 2008 Jun;134(1-2):19-38. doi: 10.1016/j.virusres.2007.12.017. Epub 2008 Feb 14. Virus Res. 2008. PMID: 18279992 Review. - Quantitative analysis of gene expression by ion-pair high-performance liquid chromatography.
Doris PA, Oefner PJ, Chilton BS, Hayward-Lester A. Doris PA, et al. J Chromatogr A. 1998 May 8;806(1):47-60. doi: 10.1016/s0021-9673(97)00514-1. J Chromatogr A. 1998. PMID: 9639880 Review.
Cited by
- Quantification of mRNA in single cells and modelling of RT-qPCR induced noise.
Bengtsson M, Hemberg M, Rorsman P, Ståhlberg A. Bengtsson M, et al. BMC Mol Biol. 2008 Jul 17;9:63. doi: 10.1186/1471-2199-9-63. BMC Mol Biol. 2008. PMID: 18631407 Free PMC article. - Development of robust isothermal RNA amplification assay for lab-free testing of RNA viruses.
Biyani R, Sharma K, Kojima K, Biyani M, Sharma V, Kumawat T, Juma KM, Yanagihara I, Fujiwara S, Kodama E, Takamura Y, Takagi M, Yasukawa K, Biyani M. Biyani R, et al. Sci Rep. 2021 Aug 6;11(1):15997. doi: 10.1038/s41598-021-95411-x. Sci Rep. 2021. PMID: 34362977 Free PMC article. - PCR inhibition by reverse transcriptase leads to an overestimation of amplification efficiency.
Suslov O, Steindler DA. Suslov O, et al. Nucleic Acids Res. 2005 Nov 27;33(20):e181. doi: 10.1093/nar/gni176. Nucleic Acids Res. 2005. PMID: 16314311 Free PMC article. - A Single Amino Acid Change to Taq DNA Polymerase Enables Faster PCR, Reverse Transcription and Strand-Displacement.
Barnes WM, Zhang Z, Kermekchiev MB. Barnes WM, et al. Front Bioeng Biotechnol. 2021 Jan 14;8:553474. doi: 10.3389/fbioe.2020.553474. eCollection 2020. Front Bioeng Biotechnol. 2021. PMID: 33520948 Free PMC article. - Quantitative analysis of FJ 194940.1 gene expression in colon cancer and its association with clinicopathological parameters.
Bartczak-Tomczyk M, Sałagacka A, Mirowski M, Jeleń A, Balcerczak E. Bartczak-Tomczyk M, et al. Contemp Oncol (Pozn). 2013;17(1):45-50. doi: 10.5114/wo.2013.33773. Epub 2013 Mar 15. Contemp Oncol (Pozn). 2013. PMID: 23788961 Free PMC article.
References
- Aatsinki J T, Lakkakorpi J T, Pietila E M, Rajaniemi H J. A coupled one-step reverse transcription PCR procedure for generation of full-length open reading frames. BioTechniques. 1994;16:282–288. - PubMed
- Alberts B, Sternglanz R. Recent excitement in the DNA replication problem. Nature. 1977;269:655–661. - PubMed
- Becker-André M. Absolute levels of mRNA by polymerase chain reaction-aided transcript titration assay. Methods Enzymol. 1993;218:420–445. - PubMed
- Becker-André M. Evaluation of absolute mRNA levels by polymerase chain reaction-aided transcript titration assay (PATTY) In: Larrick J W, Siebert P D, editors. Reverse transcriptase PCR. New York, N.Y: Ellis Horwood; 1995. pp. 121–149.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources