In situ gene expression in mixed-culture biofilms: evidence of metabolic interactions between community members - PubMed (original) (raw)
In situ gene expression in mixed-culture biofilms: evidence of metabolic interactions between community members
S Møller et al. Appl Environ Microbiol. 1998 Feb.
Abstract
Microbial communities growing in laboratory-based flow chambers were investigated in order to study compartmentalization of specific gene expression. Among the community members studied, the focus was in particular on Pseudomonas putida and a strain of an Acinetobacter sp., and the genes studied are involved in the biodegradation of toluene and related aromatic compounds. The upper-pathway promoter (Pu) and the meta-pathway promoter (Pm) from the TOL plasmid were fused independently to the gene coding for the green fluorescent protein (GFP), and expression from these promoters was studied in P. putida, which was a dominant community member. Biofilms were cultured in flow chambers, which in combination with scanning confocal laser microscopy allowed direct monitoring of promoter activity with single-cell spatial resolution. Expression from the Pu promoter was homogeneously induced by benzyl alcohol in both community and pure-culture biofilms, while the Pm promoter was induced in the mixed community but not in a pure-culture biofilm. By sequentially adding community members, induction of Pm was shown to be a consequence of direct metabolic interactions between an Acinetobacter species and P. putida. Furthermore, in fixed biofilm samples organism identity was determined and gene expression was visualized at the same time by combining GFP expression with in situ hybridization with fluorescence-labeled 16S rRNA targeting probes. This combination of techniques is a powerful approach for investigating structure-function relationships in microbial communities.
Figures
FIG. 1
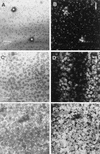
Online monitoring of Pu and Pm expression in pure-culture biofilms after 1 day of biofilm formation. (A and B) Biofilms formed by P. putida R1 (Pu::gfp). All cells are shown in the reflection image in panel A, and the GFP fluorescence in panel B shows that Pu expression was homogeneous. (C and D) Biofilms formed by P. putida R1 (Pm::gfp). All cells are shown in the reflection image in panel C, and the GFP fluorescence in panel D shows that there was no GFP expression except for that of a few bright cells, showing that Pm was not induced in the pure-culture biofilm. (E and F) Induction of Pm by 3MB. P. putida R1 (Pm::gfp) was grown for 1 day on benzyl alcohol, and then 5 mM 3MB was added to the medium. The reflection image (E) recorded on day 2 shows all cells, and the GFP signal from the same cells (F) reveals induction of Pm. The arrows indicate the direction of flow.
FIG. 2
Online monitoring of GFP expression from Pu and Pm in communities after 1 day of biofilm formation. (A and B) Pu expression in the biofilm formed by the community containing P. putida R1 (Pu::gfp). Note the spherical Acinetobacter sp. strain C6 colony (indicated by an asterisk) in the reflection image (A). The GFP fluorescence (B), visualized as an SFP, shows that Pu was constitutively expressed in the majority of the cells. (C through E) Expression of Pm in the biofilm formed by the community containing P. putida R1 (Pm::gfp). All cells (including the Acinetobacter sp. strain C6 colony indicated with an asterisk) are shown in the reflection image (C), and GFP fluorescence (D) shows the lack of homogeneous Pm expression on day 1. Panel E shows GFP expression quantified along the line shown in panel D and shows that GFP fluorescence increased as cells got closer to the Acinetobacter sp. strain C6 microcolony. The pixel intensities were measured from the maximum projection image (data not shown). (F and G) Biofilm formation by the wild-type community. The reflection image (F) and the SFP (G) show that no cells expressed GFP in the negative control. The asterisk indicates the spherical microcolonies of Acinetobacter sp. strain C6, and the arrows show the direction of flow.
FIG. 3
Addition of Acinetobacter sp. strain C6 to a P. putida R1 (Pm::gfp) biofilm. All cells are shown in the reflection image (A), and the GFP expression (B) shows that Pm was induced by the presence of Acinetobacter sp. strain C6. The asterisk indicates a spherical colony of Acinetobacter sp. strain C6, and the arrow indicates the direction of flow.
FIG. 4
Time course of GFP expression by P. putida R1 (Pm::gfp) in the community. (A and B) Lack of homogeneous expression of Pm::gfp on day 1. All cells are shown in the reflection image (A), and an optical section obtained 5 μm from the substratum shows the GFP expression (B). The asterisks indicate spherical microcolonies of Acinetobacter sp. strain C6. (C and D) Pm expression patterns on day 2. The reflection image (C) and an optical section of GFP expression obtained 5 μm from the substratum (D) show that induction occurred in bands parallel to the direction of flow. (E and F) Pm expression on day 3. The reflection image (E) shows all of the cells, and an optical section of the GFP fluorescence obtained 5 μm from the substratum (F) shows more homogeneous induction of Pm on day 3, indicating that the inducing agent accumulated in the biofilm. The arrows indicate the direction of flow.
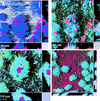
FIG. 5
Spatial distribution of organisms and gene expression in the community. The dominant organisms in the community were targeted by hybridization. (A) Dual hybridization of Acinetobacter sp. strain C6 (red) and P. putida R1 (blue) in the community containing P. putida R1 (Pu::gfp) on day 3. (B) Overlay of the GFP fluorescence (green) expressed from the Pu promoter. Cells that stained both blue (hybridization) and green (GFP expression) appear cyan. (C and D) Expression from Pm in the community containing P. putida R1 (Pm::gfp) grown for 3 days. Panel C shows a colony of Acinetobacter sp. strain C6 (red) surrounded by P. putida R1 (Pm::gfp) cells expressing GFP (cyan), and panel D shows surface coverage by Acinetobacter sp. strain C6 (red), with P. putida R1 distributed over the surface. These colonies expressed GFP (cyan). P. putida R1 was hybridized with PP986 labeled with CY5, Acinetobacter sp. strain C6 was hybridized with ACN449 labeled with CY3, and GFP was sampled in the green channel. The images shown are SFPs with x and z projections shown on the sides of the images; these projections provide extended focus images for the regions between the marks. The arrows indicate the direction of flow.
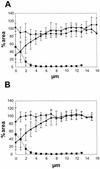
FIG. 6
Profiles of organisms and gene expression in the biofilm. (A) Expression from the Pu promoter. Symbols: ⧫, percentage of the P. putida R1 population in the biofilm expressing Pu::gfp; ▴, percentage of cells in the biofilm that were P. putida R1 cells; ▪, percentage of cells that were Acinetobacter sp. strain C6 cells. (B) Expression from the Pm promoter. Symbols: ⧫, percentage of the P. putida R1 population in the biofilm expressing Pm::gfp; ▴, percentage of cells in the biofilm that were P. putida R1 cells; ▪, percentage of cells that were Acinetobacter sp. strain C6 cells. Percentages were calculated by determining the ratio between areas covered by the respective fluorescent signals in a series of multicolor optical sections. All bacteria were hybridized by using EUB338 labeled with CY5, P. putida R1 was hybridized with PP986 labeled with CY3 or CY5, Acinetobacter sp. strain C6 was hybridized with ACN449 labeled with CY3, and GFP was sampled in the green channel. Error bars indicate ±1 standard deviation. The profiles are the results obtained after we averaged the data for five independent positions in the biofilm.
Similar articles
- Establishment of new genetic traits in a microbial biofilm community.
Christensen BB, Sternberg C, Andersen JB, Eberl L, Moller S, Givskov M, Molin S. Christensen BB, et al. Appl Environ Microbiol. 1998 Jun;64(6):2247-55. doi: 10.1128/AEM.64.6.2247-2255.1998. Appl Environ Microbiol. 1998. PMID: 9603843 Free PMC article. - Metabolic commensalism and competition in a two-species microbial consortium.
Christensen BB, Haagensen JA, Heydorn A, Molin S. Christensen BB, et al. Appl Environ Microbiol. 2002 May;68(5):2495-502. doi: 10.1128/AEM.68.5.2495-2502.2002. Appl Environ Microbiol. 2002. PMID: 11976126 Free PMC article. - Distribution of bacterial growth activity in flow-chamber biofilms.
Sternberg C, Christensen BB, Johansen T, Toftgaard Nielsen A, Andersen JB, Givskov M, Molin S. Sternberg C, et al. Appl Environ Microbiol. 1999 Sep;65(9):4108-17. doi: 10.1128/AEM.65.9.4108-4117.1999. Appl Environ Microbiol. 1999. PMID: 10473423 Free PMC article. - Transcriptional control of the Pseudomonas putida TOL plasmid catabolic pathways.
Marqués S, Ramos JL. Marqués S, et al. Mol Microbiol. 1993 Sep;9(5):923-9. doi: 10.1111/j.1365-2958.1993.tb01222.x. Mol Microbiol. 1993. PMID: 7934920 Review. - Microbial communities and their interactions in biofilm systems: an overview.
Wuertz S, Okabe S, Hausner M. Wuertz S, et al. Water Sci Technol. 2004;49(11-12):327-36. Water Sci Technol. 2004. PMID: 15303758 Review.
Cited by
- Cryosectioning yeast communities for examining fluorescence patterns.
Momeni B, Shou W. Momeni B, et al. J Vis Exp. 2012 Dec 26;(70):50101. doi: 10.3791/50101. J Vis Exp. 2012. PMID: 23287845 Free PMC article. - Expression of green fluorescent protein in Streptococcus gordonii DL1 and its use as a species-specific marker in coadhesion with Streptococcus oralis 34 in saliva-conditioned biofilms in vitro.
Aspiras MB, Kazmerzak KM, Kolenbrander PE, McNab R, Hardegen N, Jenkinson HF. Aspiras MB, et al. Appl Environ Microbiol. 2000 Sep;66(9):4074-83. doi: 10.1128/AEM.66.9.4074-4083.2000. Appl Environ Microbiol. 2000. PMID: 10966431 Free PMC article. - Ecological advantages of autolysis during the development and dispersal of Pseudoalteromonas tunicata biofilms.
Mai-Prochnow A, Webb JS, Ferrari BC, Kjelleberg S. Mai-Prochnow A, et al. Appl Environ Microbiol. 2006 Aug;72(8):5414-20. doi: 10.1128/AEM.00546-06. Appl Environ Microbiol. 2006. PMID: 16885293 Free PMC article. - Bacterial biofilm within diseased pancreatic and biliary tracts.
Swidsinski A, Schlien P, Pernthaler A, Gottschalk U, Bärlehner E, Decker G, Swidsinski S, Strassburg J, Loening-Baucke V, Hoffmann U, Seehofer D, Hale LP, Lochs H. Swidsinski A, et al. Gut. 2005 Mar;54(3):388-95. doi: 10.1136/gut.2004.043059. Gut. 2005. PMID: 15710988 Free PMC article. - Biofilm tolerance, resistance and infections increasing threat of public health.
Yang S, Li X, Cang W, Mu D, Ji S, An Y, Wu R, Wu J. Yang S, et al. Microb Cell. 2023 Sep 26;10(11):233-247. doi: 10.15698/mic2023.11.807. eCollection 2023 Nov 6. Microb Cell. 2023. PMID: 37933277 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases