Intranuclear targeting of AML/CBFalpha regulatory factors to nuclear matrix-associated transcriptional domains - PubMed (original) (raw)
. 1998 Feb 17;95(4):1585-9.
doi: 10.1073/pnas.95.4.1585.
S McNeil, S Pockwinse, J Nickerson, L Shopland, J B Lawrence, S Penman, S Hiebert, J B Lian, A J van Wijnen, J L Stein, G S Stein
Affiliations
- PMID: 9465059
- PMCID: PMC19104
- DOI: 10.1073/pnas.95.4.1585
Intranuclear targeting of AML/CBFalpha regulatory factors to nuclear matrix-associated transcriptional domains
C Zeng et al. Proc Natl Acad Sci U S A. 1998.
Abstract
The AML/CBFalpha runt transcription factors are key regulators of hematopoietic and bone tissue-specific gene expression. These factors contain a 31-amino acid nuclear matrix targeting signal that supports association with the nuclear matrix. We determined that the AML/CBFalpha factors must bind to the nuclear matrix to exert control of transcription. Fusing the nuclear matrix targeting signal to the GAL4 DNA binding domain transactivates a genomically integrated GAL4 responsive reporter gene. These data suggest that AML/CBFalpha must associate with the nuclear matrix to effect transcription. We used fluorescence labeling of epitope-tagged AML-1B (CBFA2) to show it colocalizes with a subset of hyperphosphorylated RNA polymerase II molecules concentrated in foci and linked to the nuclear matrix. This association of AML-1B with RNA polymerase II requires active transcription and a functional DNA binding domain. The nuclear matrix domains that contain AML-1B are distinct from SC35 RNA processing domains. Our results suggest two of the requirements for AML-dependent transcription initiation by RNA polymerase II are association of AML-1B with the nuclear matrix together with specific binding of AML to gene promoters.
Figures
Figure 1
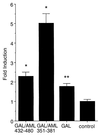
The NMTS of AML-1B trans-activates heterologous reporter gene expression. The reporter gene is chromosomally integrated and is composed of multimerized GAL4-sites fused to the Adenovirus minimal E1A promoter that drives expression of luciferase activity. Data are expressed as fold-induction in response to CMV driven coexpression of fusion-proteins between the GAL4 DNA binding domain (aa 1–147; GAL) and C-terminal segments of AML-1B (GAL/AML 432–480 and GAL/AML 351–381), relative to cells transfected with the CMV-vector alone (control). Each bar represents the data from 5 experiments (2–3 replicates). The asterisks indicates statistically significant differences relative to the control (*P ≤ 0.0005; **P ≤ 0.005).
Figure 2
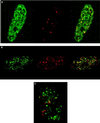
AML-1B is targeted to transcriptionally active nuclear matrix domains. (A and B) A subset of AML-1B and RNA polymerase II are colocalized in the nuclear matrix. Human osteosarcoma SAOS-2 cells were transiently transfected with a construct expressing HA-tagged CBFα2/AML-1B. In situ nuclear matrix samples were prepared 24 hr after transfection as described in Materials and Methods. HA/AML-1B was detected with an antibody against HA (green) (Left) and RNA polymerase II0 with the B3 anti-RNA polymerase II antibody (red) (Center). The merged image is shown in the Right panel; yellow indicates colocalization of AML-1B with RNA polymerase II0. The colocalization of AML-1B with RNA polymerase II0 was evaluated by digital immunofluorescence microscopy (A) and by confocal microscopy (B) with a series of optical sections (0.5 μm intervals) through a single cell. B shows single-color and merged images (green, HA/AML-1B; red, RNA polymerase II; yellow, colocalization) for one of the optical sections. (C) Promoter recognition is required for the colocalization of AML-1B with RNA polymerase II. SAOS-2 cells were transfected with the HA-tagged L148D mutant that contains a point mutation in the runt homology domain and lacks DNA-binding activity. In situ nuclear matrix preparations were visualized as described in A (green, HA/AML-1B; red, RNA polymerase II; yellow, colocalization). The images shown were obtained by digital fluorescence microscopy. Similar images were obtained by directly photographing immunofluorescence signals (data not shown).
Figure 3
AML-1B/RNA polymerase II0 sites are coupled with RNA synthesis. The HA/AML-1B construct was transfected into SAOS and ROS 17/2.8 cells. BrUTP labeling was performed with Triton X-100 permeabilized cells. Cells were double-stained with the B3 anti RNA polymerase II antibody and the BU33 anti-BrdUrd antibody. BrUTP (green) and RNA polymerase II0 (red) single-color channels and the merged image are each shown, with yellow indicating colocalization of BrUTP and RNA polymerase II0. The same extent of colocalization was observed in mock-transfected and untransfected cells (data not shown).
Figure 4
AML-1B does not colocalize with SC-35 RNA processing domain. SAOS cells were transfected with HA/AML-1B and evaluated by immunofluorescence analysis of the nuclear matrix in situ by using the HA antibody (green, AML-1B) and the SC-35 antibody (red, SC-35) that recognizes SC-35 RNA processing domain. In some cells, proximity of AML-1B and SC-35 domains is reflected by limited yellow staining.
Similar articles
- Identification of a nuclear matrix targeting signal in the leukemia and bone-related AML/CBF-alpha transcription factors.
Zeng C, van Wijnen AJ, Stein JL, Meyers S, Sun W, Shopland L, Lawrence JB, Penman S, Lian JB, Stein GS, Hiebert SW. Zeng C, et al. Proc Natl Acad Sci U S A. 1997 Jun 24;94(13):6746-51. doi: 10.1073/pnas.94.13.6746. Proc Natl Acad Sci U S A. 1997. PMID: 9192636 Free PMC article. - Targeting of the YY1 transcription factor to the nucleolus and the nuclear matrix in situ: the C-terminus is a principal determinant for nuclear trafficking.
McNeil S, Guo B, Stein JL, Lian JB, Bushmeyer S, Seto E, Atchison ML, Penman S, van Wijnen AJ, Stein GS. McNeil S, et al. J Cell Biochem. 1998 Mar 15;68(4):500-10. J Cell Biochem. 1998. PMID: 9493912 - Bone tissue specific transcriptional control: options for targeting gene therapy to the skeleton.
Stein GS, Lian JB, Stein JL, van Wijnen AJ. Stein GS, et al. Cancer. 2000 Jun 15;88(12 Suppl):2899-902. doi: 10.1002/1097-0142(20000615)88:12+<2899::aid-cncr3>3.0.co;2-o. Cancer. 2000. PMID: 10898331 Review. - Intranuclear trafficking of transcription factors: implications for biological control.
Stein GS, van Wijnen AJ, Stein JL, Lian JB, Montecino M, Choi J, Zaidi K, Javed A. Stein GS, et al. J Cell Sci. 2000 Jul;113 ( Pt 14):2527-33. doi: 10.1242/jcs.113.14.2527. J Cell Sci. 2000. PMID: 10862710 Review.
Cited by
- Integration of Runx and Smad regulatory signals at transcriptionally active subnuclear sites.
Zaidi SK, Sullivan AJ, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Zaidi SK, et al. Proc Natl Acad Sci U S A. 2002 Jun 11;99(12):8048-53. doi: 10.1073/pnas.112664499. Proc Natl Acad Sci U S A. 2002. PMID: 12060751 Free PMC article. - Altered localization of retinoid X receptor alpha coincides with loss of retinoid responsiveness in human breast cancer MDA-MB-231 cells.
Tanaka T, Dancheck BL, Trifiletti LC, Birnkrant RE, Taylor BJ, Garfield SH, Thorgeirsson U, De Luca LM. Tanaka T, et al. Mol Cell Biol. 2004 May;24(9):3972-82. doi: 10.1128/MCB.24.9.3972-3982.2004. Mol Cell Biol. 2004. PMID: 15082790 Free PMC article. - Multiple subnuclear targeting signals of the leukemia-related AML1/ETO and ETO repressor proteins.
Barseguian K, Lutterbach B, Hiebert SW, Nickerson J, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Barseguian K, et al. Proc Natl Acad Sci U S A. 2002 Nov 26;99(24):15434-9. doi: 10.1073/pnas.242588499. Epub 2002 Nov 11. Proc Natl Acad Sci U S A. 2002. PMID: 12427969 Free PMC article. - RUNX1 regulates the CD34 gene in haematopoietic stem cells by mediating interactions with a distal regulatory element.
Levantini E, Lee S, Radomska HS, Hetherington CJ, Alberich-Jorda M, Amabile G, Zhang P, Gonzalez DA, Zhang J, Basseres DS, Wilson NK, Koschmieder S, Huang G, Zhang DE, Ebralidze AK, Bonifer C, Okuno Y, Gottgens B, Tenen DG. Levantini E, et al. EMBO J. 2011 Aug 26;30(19):4059-70. doi: 10.1038/emboj.2011.285. EMBO J. 2011. PMID: 21873977 Free PMC article. - An architectural perspective of vitamin D responsiveness.
Montecino M, Stein GS, Cruzat F, Marcellini S, Stein JL, Lian JB, van Wijnen AJ, Arriagada G. Montecino M, et al. Arch Biochem Biophys. 2007 Apr 15;460(2):293-9. doi: 10.1016/j.abb.2006.12.015. Epub 2007 Jan 8. Arch Biochem Biophys. 2007. PMID: 17288986 Free PMC article. Review.
References
- Nickerson J A, Blencowe B J, Penman S. In: The Architectural Organization of Nuclear Metabolism. Berezney R, Jeon K W, editors. 162A. NY: Academic Press; 1995. pp. 67–123. - PubMed
- Stein G S, Stein J L, Lian J B, van Wijnen A J, Montecino M. J Cell Biochem. 1996;62:198–209. - PubMed
- Berezney R, Mortillaro M, Ma H, Meng C, Samarabandu J, Wei X, Somanathan S, Liou W S, Pan S J, Cheng P C. J Cell Biochem. 1996;62:223–226. - PubMed
- Jackson D A. Mol Biol Rep. 1997;24:209–220. - PubMed
- Davie J R. Mol Biol Rep. 1997;24:197–207. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources