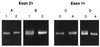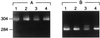Conformation sensitive gel electrophoresis for simple and accurate detection of mutations: comparison with denaturing gradient gel electrophoresis and nucleotide sequencing - PubMed (original) (raw)
Comparative Study
Conformation sensitive gel electrophoresis for simple and accurate detection of mutations: comparison with denaturing gradient gel electrophoresis and nucleotide sequencing
J Körkkö et al. Proc Natl Acad Sci U S A. 1998.
Abstract
Previously, an assay called conformation sensitive gel electrophoresis (CSGE) was developed for scanning PCR products for the presence of single-base and larger base mismatches in DNA. The assay was based on the assumption that mildly denaturing solvents in an appropriate buffer can accentuate the conformational changes produced by single-base mismatches in double-stranded DNA and thereby increase the differential migration in electrophoretic gels of heteroduplexes and homoduplexes. Here the sensitivity of assays by CSGE was improved by limiting the maximal size of the PCR products to 450 bp and making several changes in the conditions for PAGE. With the improved conditions, CSGE detected all 76 previously identified single-base changes in a large series of PCR products from collagen genes that contain multiple exons with highly repetitive and GC-rich sequences. In a survey of 736 alleles of collagen genes, CSGE detected 223 unique single-base mismatches that were confirmed by nucleotide sequencing. CSGE has the advantage over other methods for scanning PCR products in that it is simple, requires no special preparation of PCR products, has a large capacity, and does not use radioactivity.
Figures
Figure 1
The effect of product size on detection of sequence variations by CSGE. (Upper) CSGE analysis of a 755-bp PCR product that contains sequences for exons 25 and 26 of the COL1A1 gene. No heteroduplexes are detected. (Lower) CSGE analysis of a 276-bp PCR product that contains sequences for exon 25 of the same gene. DNA from the same five individuals were analyzed in both panels. Heteroduplexes were detected in samples 3 and 5.
Figure 2
(A and B) Effect of electrophoretic conditions on separation of heteroduplexes from PCR products of exon 21 from the COL1A1 gene. (A) Separation on a 10% polyacrylamide gel at 400 V for 18 hr. (B) Same samples as in A separated on a 10% polyacrylamide gel at 40 W for 6 hr. Sample 1 has a polymorphism. (C) Electrophoresis in a 10% polyacrylamide gel at 40 W for 6 hr. (D) Same samples as in C separated in a 15% polyacrylamide gel at 40 W for 8.5 hr. Sample 3 has a polymorphism.
Figure 3
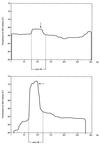
Melting profile for a PCR product containing sequences for exon 28 of the COL3A1 gene. (Upper) Melting profile after addition of 10-bp GC clamps to the 5′-end and 3′-end of the product. (Lower) Melting profile for the native sequence without GC clamps. Arrows indicate the site of a single-base mismatch.
Figure 4
CSGE analysis of the PCR products for exon 28 of the COL3A1 gene. (A) PCR samples with 10-bp GC clamps. (B) Same PCR product without GC clamps. Heteroduplexes caused by a C to T polymorphism in sample 3 are seen only in B.
Figure 5
Detection of sequence variations in high-melting domains in exon 30 (C to A) and exon 31 (G to A) of the COL3A1 gene, and exon 5B (A to C) of the COL2A1 gene. Previously not detected or poorly detected sequence variations (22) were detected here in samples 1, 3, and 5 by using 15% polyacrylamide gels and 40 W/8.5 hr. Samples 2, 4, and 6 are controls.
Figure 6
Detection of more than one base mismatch in PCR products for exon 30 of the COL1A1 gene. The PCR products of 236 bp each were obtained from seven unrelated individuals. For clarity, the mismatches are defined here as A (C−5IVS29A), B (T39IVS30C), C (A−41IVS29G), and D (G−43IVS29C). Lane 1, mismatches A and B. Lane 2, mismatches A, B, and C. Lane 3, mismatches C and D. Lane 4, mismatch B. Lane 5, mismatches B and C. Lane 6, mismatch C. Lane 7, control sample.
Similar articles
- Conformation-sensitive gel electrophoresis for rapid detection of single-base differences in double-stranded PCR products and DNA fragments: evidence for solvent-induced bends in DNA heteroduplexes.
Ganguly A, Rock MJ, Prockop DJ. Ganguly A, et al. Proc Natl Acad Sci U S A. 1993 Nov 1;90(21):10325-9. doi: 10.1073/pnas.90.21.10325. Proc Natl Acad Sci U S A. 1993. PMID: 8234293 Free PMC article. - An update on conformation sensitive gel electrophoresis.
Ganguly A. Ganguly A. Hum Mutat. 2002 Apr;19(4):334-42. doi: 10.1002/humu.10059. Hum Mutat. 2002. PMID: 11933188 Review. - [Application studies on the gene diagnosis and carrier detection of hemophilia A by using polymerase chain reaction-conformation sensitive gel electrophoresis].
Lillicrap D, He GP, Leggo J, Liu YS, Tong XH, Zhou GX, Luo LH. Lillicrap D, et al. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2009 Aug;26(4):393-9. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2009. PMID: 20017302 Chinese. - Mutation detection by denaturing gradient gel electrophoresis (DGGE).
Fodde R, Losekoot M. Fodde R, et al. Hum Mutat. 1994;3(2):83-94. doi: 10.1002/humu.1380030202. Hum Mutat. 1994. PMID: 8199599 Review.
Cited by
- Mitochondrial DNA variant m.15218A > G in Finnish epilepsy patients who have maternal relatives with epilepsy, sensorineural hearing impairment or diabetes mellitus.
Soini HK, Moilanen JS, Vilmi-Kerälä T, Finnilä S, Majamaa K. Soini HK, et al. BMC Med Genet. 2013 Jul 19;14:73. doi: 10.1186/1471-2350-14-73. BMC Med Genet. 2013. PMID: 23870133 Free PMC article. - Analysis of the RNASEL gene in familial and sporadic prostate cancer.
Wang L, McDonnell SK, Elkins DA, Slager SL, Christensen E, Marks AF, Cunningham JM, Peterson BJ, Jacobsen SJ, Cerhan JR, Blute ML, Schaid DJ, Thibodeau SN. Wang L, et al. Am J Hum Genet. 2002 Jul;71(1):116-23. doi: 10.1086/341281. Epub 2002 May 17. Am J Hum Genet. 2002. PMID: 12022038 Free PMC article. - Mutation analysis of the CHK2 gene in families with hereditary breast cancer.
Allinen M, Huusko P, Mäntyniemi S, Launonen V, Winqvist R. Allinen M, et al. Br J Cancer. 2001 Jul 20;85(2):209-12. doi: 10.1054/bjoc.2001.1858. Br J Cancer. 2001. PMID: 11461078 Free PMC article. - The mitochondrial gene tree comes of age.
Richards M, Macaulay V. Richards M, et al. Am J Hum Genet. 2001 Jun;68(6):1315-20. doi: 10.1086/320615. Epub 2001 May 10. Am J Hum Genet. 2001. PMID: 11349234 Free PMC article. No abstract available. - Defective C-propeptides of the proalpha2(I) chain of type I procollagen impede molecular assembly and result in osteogenesis imperfecta.
Pace JM, Wiese M, Drenguis AS, Kuznetsova N, Leikin S, Schwarze U, Chen D, Mooney SH, Unger S, Byers PH. Pace JM, et al. J Biol Chem. 2008 Jun 6;283(23):16061-7. doi: 10.1074/jbc.M801982200. Epub 2008 Mar 27. J Biol Chem. 2008. PMID: 18375391 Free PMC article.
References
- Kuivaniemi H, Tromp G, Prockop D J. Hum Mutat. 1997;9:300–315. - PubMed
- Cotton R G H. Mutat Res. 1993;285:125–144. - PubMed
- Grompe M. Nat Genet. 1993;5:111–117. - PubMed
- Glavac D, Dean M. Hum Mutat. 1995;6:281–287. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous