Phosphate availability affects the thylakoid lipid composition and the expression of SQD1, a gene required for sulfolipid biosynthesis in Arabidopsis thaliana - PubMed (original) (raw)
Phosphate availability affects the thylakoid lipid composition and the expression of SQD1, a gene required for sulfolipid biosynthesis in Arabidopsis thaliana
B Essigmann et al. Proc Natl Acad Sci U S A. 1998.
Abstract
Photosynthetic membranes of higher plants contain specific nonphosphorous lipids like the sulfolipid sulfoquinovosyl diacylglycerol in addition to the ubiquitous phospholipid phosphatidylglycerol. In bacteria, an environmental factor that drastically affects thylakoid lipid composition appears to be the availability of phosphate. Accordingly, we discovered an increase in the relative amount of sulfolipid and a concomitant decrease in phosphatidylglycerol in Arabidopsis thaliana grown on medium with reduced amounts of phosphate, as well as in the pho1 mutant of A. thaliana deficient in phosphate transport. To investigate the molecular basis of the observed change in lipid composition, we isolated a cDNA of A. thaliana, designated SQD1, that encodes a protein involved in sulfolipid biosynthesis as suggested by three lines of evidence. First, the cDNA shows high sequence similarity to bacterial sqdB genes known to be essential for sulfolipid biosynthesis; second, the SQD1 gene product is imported into chloroplasts where sulfolipid biosynthesis takes place; and third, transgenic plants expressing SQD1 in antisense orientation show a reduction in sulfolipid content. In the pho1 mutant as well as in wild-type plants grown under reduced phosphate availability, increased amounts of SQD1 mRNA and SQD1 protein are detected, suggesting that the increase in sulfolipid content under phosphate limitation is the result of an increased expression of at least one gene required for sulfolipid biosynthesis in A. thaliana. It is suggested that a certain amount of anionic thylakoid lipid is maintained by substituting sulfolipid for phosphatidylglycerol under reduced phosphate availability.
Figures
Figure 1
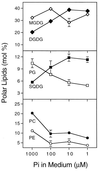
Changes in relative amounts of complex lipids of wild-type A. thaliana in response to changing phosphate concentration. The plants were germinated and grown for 15 days on agar-solidified mineral medium as described under methods. Values represent the means ± SD of at least three independent measurements. Error bars are indicated, if they are not smaller than the symbol size. DGDG, digalactosyl diacylglycerol; MGDG, monogalactosyl diacylglycerol; PC, phosphatidylcholine; PE, phosphatidyl ethanolamine; PG, phosphatidylglycerol; SQDG, sulfoquinovosyl diacylglycerol.
Figure 2
Comparison of the amino acid sequences predicted from the sqdB gene of the cyanobacterium Synechococcus sp. PCC7942 (Syne) and the SQD1 cDNA of A. thaliana (Arab). Identical amino acid residues are marked by black boxes.
Figure 3
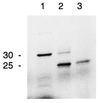
Uptake and processing of a carboxy-terminally truncated SQD1 preprotein of A. thaliana by isolated pea chloroplasts. Lane 1, translation product; lane 2, translation product incubated in the presence of pea chloroplasts; lane 3, translation product incubated in the presence of pea chloroplasts and thermolysin. Proteins were labeled with [35S]methionine and visualized by autoradiography after SDS/PAGE. The approximate molecular mass (kDa) of the proteins is indicated.
Figure 4
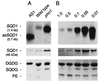
Expression of the SQD1 gene in leaves of A. thaliana. (A) Wild type, the strongest SQD1 antisense line AS1, and the pho1 mutant grown for 14 days in soil. (B) Wild type germinated and raised for 7 days on agar-solidified Murashige–Skoog medium and subsequently grown for 7 days on mineral medium containing a concentration of Pi (mM) as indicated. (Upper) SQD1 mRNA (SQD1) or the presumed antisense RNA (asSQD1) detected by hybridization with the SQD1 cDNA (the approximate sizes are indicated). (Middle) SQD1 protein (SQD1) detected by immunoblotting (the apparent molecular mass is indicated). (Lower) Central part of iodine-stained lipid chromatograms (DGDG, digalactosyl diacylglycerol; PE, phosphatidylethanolamine; SQDG, sulfoquinovosyl diacylglycerol). RNA, protein, and lipids were isolated from identical batches of plants. Equal amounts of total RNA, protein, or lipids were loaded based on visual examination of stained gels or TLC plates.
Similar articles
- Prediction of the active-site structure and NAD(+) binding in SQD1, a protein essential for sulfolipid biosynthesis in Arabidopsis.
Essigmann B, Hespenheide BM, Kuhn LA, Benning C. Essigmann B, et al. Arch Biochem Biophys. 1999 Sep 1;369(1):30-41. doi: 10.1006/abbi.1999.1344. Arch Biochem Biophys. 1999. PMID: 10462438 - Arabidopsis disrupted in SQD2 encoding sulfolipid synthase is impaired in phosphate-limited growth.
Yu B, Xu C, Benning C. Yu B, et al. Proc Natl Acad Sci U S A. 2002 Apr 16;99(8):5732-7. doi: 10.1073/pnas.082696499. Proc Natl Acad Sci U S A. 2002. PMID: 11960029 Free PMC article. - Disruption of a gene essential for sulfoquinovosyldiacylglycerol biosynthesis in Sinorhizobium meliloti has no detectable effect on root nodule symbiosis.
Weissenmayer B, Geiger O, Benning C. Weissenmayer B, et al. Mol Plant Microbe Interact. 2000 Jun;13(6):666-72. doi: 10.1094/MPMI.2000.13.6.666. Mol Plant Microbe Interact. 2000. PMID: 10830266 - Biosynthesis and functions of the plant sulfolipid.
Shimojima M. Shimojima M. Prog Lipid Res. 2011 Jul;50(3):234-9. doi: 10.1016/j.plipres.2011.02.003. Epub 2011 Mar 1. Prog Lipid Res. 2011. PMID: 21371504 Review. - Three enzyme systems for galactoglycerolipid biosynthesis are coordinately regulated in plants.
Benning C, Ohta H. Benning C, et al. J Biol Chem. 2005 Jan 28;280(4):2397-400. doi: 10.1074/jbc.R400032200. Epub 2004 Dec 6. J Biol Chem. 2005. PMID: 15590685 Review.
Cited by
- Evidence for phosphate-dependent control of symbiont cell division in the model anemone Exaiptasia diaphana.
Faulstich NG, Deloach AR, Ksor YB, Mesa GH, Sharma DS, Sisk SL, Mitchell GC. Faulstich NG, et al. mBio. 2024 Sep 11;15(9):e0105924. doi: 10.1128/mbio.01059-24. Epub 2024 Aug 6. mBio. 2024. PMID: 39105583 Free PMC article. - A multi-omic survey of black cottonwood tissues highlights coordinated transcriptomic and metabolomic mechanisms for plant adaptation to phosphorus deficiency.
Kangi E, Brzostek ER, Bills RJ, Callister SJ, Zink EM, Kim YM, Larsen PE, Cumming JR. Kangi E, et al. Front Plant Sci. 2024 Apr 5;15:1324608. doi: 10.3389/fpls.2024.1324608. eCollection 2024. Front Plant Sci. 2024. PMID: 38645387 Free PMC article. - Structure, biogenesis, and evolution of thylakoid membranes.
Ostermeier M, Garibay-Hernández A, Holzer VJC, Schroda M, Nickelsen J. Ostermeier M, et al. Plant Cell. 2024 Oct 3;36(10):4014-4035. doi: 10.1093/plcell/koae102. Plant Cell. 2024. PMID: 38567528 Review. - Preferential phosphatidylglycerol synthesis via phosphorus supply through rRNA degradation in the cyanobacterium, Synechocystis sp. PCC 6803, under phosphate-starved conditions.
Hiyoshi T, Haga M, Sato N. Hiyoshi T, et al. Front Plant Sci. 2024 Jan 29;15:1335085. doi: 10.3389/fpls.2024.1335085. eCollection 2024. Front Plant Sci. 2024. PMID: 38348270 Free PMC article. - Making watercress (Nasturtium officinale) cropping sustainable: genomic insights into enhanced phosphorus use efficiency in an aquatic crop.
Hibbert LE, Qian Y, Smith HK, Milner S, Katz E, Kliebenstein DJ, Taylor G. Hibbert LE, et al. Front Plant Sci. 2023 Nov 7;14:1279823. doi: 10.3389/fpls.2023.1279823. eCollection 2023. Front Plant Sci. 2023. PMID: 38023842 Free PMC article.
References
- Webb M S, Green B R. Biochim Biophys Acta. 1991;1060:133–158.
- Benning C, Huang Z H, Gage D A. Arch Biochem Biophys. 1995;317:103–111. - PubMed
- Güler S, Seeliger A, Härtel H, Renger G, Benning C. J Biol Chem. 1996;271:7501–7507. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases