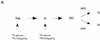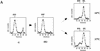The Cdc7 protein kinase is required for origin firing during S phase - PubMed (original) (raw)
The Cdc7 protein kinase is required for origin firing during S phase
K Bousset et al. Genes Dev. 1998.
Erratum in
- Genes Dev 1998 Apr 1;12(7):1072
Abstract
The Cdc7p protein kinase plays an essential, but undefined, role promoting S phase in the budding yeast, Saccharomyces cerevisiae. Previous experiments have shown that the essential function of Cdc7 is executed near the G1-S boundary; after Start but before the elongation phase of DNA replication. Origins of DNA replication fire throughout S phase in budding yeast. Therefore, the G1-S transition is a cell-cycle event that precedes, and is distinct from, the activation of individual origins. Consequently, we have asked whether Cdc7 is only required for S-phase entry or if it plays a role during S phase in origin firing. In this article, we show that partial loss of Cdc7 function results in slow progression through S phase rather than slow entry into S phase and that Cdc7 is still required for the timely completion of S phase after a block to elongation with hydroxyurea. This is because Cdc7 is still required for the activation of late-firing origins after the hydroxyurea block. These experiments show that, rather than acting as a global regulator of the G1-S transition, Cdc7 appears to play a more direct role in the firing of replication origins during S phase.
Figures
Figure 1
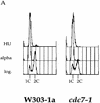
cdc7 mutants are defective in S-phase progression, not G1–S transition. (A,B) Flow cytometric analysis of either cdc7-4 (A) or the cdc7-1 (B) temperature-sensitive cells (top) and the parental wild-type strain (bottom). Cells blocked in G1 with α factor at 24°C were shifted to the indicated temperatures for 30 min and released from the block. Samples were collected every 20 min. Positions of 1C and 2C DNA contents are indicated below each histogram.
Figure 1
cdc7 mutants are defective in S-phase progression, not G1–S transition. (A,B) Flow cytometric analysis of either cdc7-4 (A) or the cdc7-1 (B) temperature-sensitive cells (top) and the parental wild-type strain (bottom). Cells blocked in G1 with α factor at 24°C were shifted to the indicated temperatures for 30 min and released from the block. Samples were collected every 20 min. Positions of 1C and 2C DNA contents are indicated below each histogram.
Figure 2
Cdc7 is important, but not essential, after a HU block. (A) Flow cytometric analysis of cdc7-1 cells (right) and the parental wild-type cells (left) is shown. Cells were grown in logarithmic phase (log.), arrested in G1 with α factor (alpha), and then released into HU for 90 min at 24°C. (B) The cells arrested with HU (A) were held in HU at either 24°C (top) or 37°C (bottom) for 30 min to inactivate Cdc7 in the cdc7-1 strain. They were then washed (0 min sample) and released from the HU block at either 24°C or 37°C, respectively. Every 15 min, aliquots were withdrawn to follow the cell-cycle progression.
Figure 2
Cdc7 is important, but not essential, after a HU block. (A) Flow cytometric analysis of cdc7-1 cells (right) and the parental wild-type cells (left) is shown. Cells were grown in logarithmic phase (log.), arrested in G1 with α factor (alpha), and then released into HU for 90 min at 24°C. (B) The cells arrested with HU (A) were held in HU at either 24°C (top) or 37°C (bottom) for 30 min to inactivate Cdc7 in the cdc7-1 strain. They were then washed (0 min sample) and released from the HU block at either 24°C or 37°C, respectively. Every 15 min, aliquots were withdrawn to follow the cell-cycle progression.
Figure 3
A plasmid containing ARS301 as sole origin of replication replicates late in S phase. Replication of ARS301 on pCS1 was followed through S phase in comparison with the early-firing origin ARS305 on chromosome III and the late-replicating sequence R11 on chromosome V by density transfer as described in Materials and Methods. Percentage of replication of pCS1 (□), ARS305 (⋄), and R11 (○) is plotted against the time after release from a cdc7 block. Dotted (shaded) lines indicate the replication times _t_rep for the three sequences of interest. See Table 1 for the times determined. _t_rep is defined as the time when half of a specific sequence has replicated (i.e., shifted to the HL peak).
Figure 4
Cdc7 is not required for replication of a plasmid containing the early replication origin ARS305 after HU. (A) Outline of the experimental procedure (see text for details). (B) RM14-3a cells (cdc7-1) containing an ARS305 plasmid (p305.2) were treated as illustrated in A. Cells were grown in heavy medium and arrested with α factor (α). They were subsequently released into HU in light medium (HU), and then released from HU at either 24°C or 37°C for 3 hr as described in Materials and Methods. DNA was isolated and analyzed for replication. Replication of ARS305 on the plasmid was analyzed by DNA blot hybridization with a plasmid-specific probe (see Materials and Methods). The relative amounts of radioactivity are plotted against the fraction numbers. Location of HH and HL DNA are indicated. Additionally, the HH peak as it appears in the G1 block is superimposed onto the other plots as a shaded line.
Figure 4
Cdc7 is not required for replication of a plasmid containing the early replication origin ARS305 after HU. (A) Outline of the experimental procedure (see text for details). (B) RM14-3a cells (cdc7-1) containing an ARS305 plasmid (p305.2) were treated as illustrated in A. Cells were grown in heavy medium and arrested with α factor (α). They were subsequently released into HU in light medium (HU), and then released from HU at either 24°C or 37°C for 3 hr as described in Materials and Methods. DNA was isolated and analyzed for replication. Replication of ARS305 on the plasmid was analyzed by DNA blot hybridization with a plasmid-specific probe (see Materials and Methods). The relative amounts of radioactivity are plotted against the fraction numbers. Location of HH and HL DNA are indicated. Additionally, the HH peak as it appears in the G1 block is superimposed onto the other plots as a shaded line.
Figure 5
Cdc7 is required for replication of a plasmid containing ARS301 after HU. RM14-3a cells (cdc7-1) containing the ARS301 plasmid (pCS1) were treated and their DNA was analyzed as described for Fig. 4. (A) Replication of a late-replicating sequence on chromosome V was analyzed by DNA blot hybridization with the R11 probe. (B) Replication of a plasmid containing ARS301 as the sole origin was analyzed by probing the same fractions with a probe specific for the vector (see Materials and Methods). The relative amounts of radioactivity is plotted against the fraction number. Location of HH and HL DNAs are indicated.
Figure 5
Cdc7 is required for replication of a plasmid containing ARS301 after HU. RM14-3a cells (cdc7-1) containing the ARS301 plasmid (pCS1) were treated and their DNA was analyzed as described for Fig. 4. (A) Replication of a late-replicating sequence on chromosome V was analyzed by DNA blot hybridization with the R11 probe. (B) Replication of a plasmid containing ARS301 as the sole origin was analyzed by probing the same fractions with a probe specific for the vector (see Materials and Methods). The relative amounts of radioactivity is plotted against the fraction number. Location of HH and HL DNAs are indicated.
Figure 6
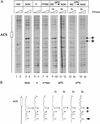
Cdc7 is required to convert plasmid-borne ARS301 to the postreplicative state after HU. (A) cdc7-1 cells (2032) containing the ARS301 plasmid (pCS1) were arrested in G2/M by nocodazole (lanes 3,4) or in G1 by α factor (lanes 5,6). G1-arrested cells were released into HU (lanes 7,8). The culture was split in two. Each half was held in HU and incubated at either 24°C or 37°C for 30 min before release into nocodazole at these temperatures. Samples were taken for genomic footprinting analysis 1 hr and 3 hr after release from HU into nocodazole (lanes 9–16). The primer (JD63b) is specific for the plasmid borne ARS301. Lanes 1 and 2 show primer extensions performed on naked DNA. Exposure levels were adjusted because plasmid recovery was lower from G1 and S-phase cells and on release from HU at 37°C. (B) Densitometric quantification of Fig. 4A, lanes 4, 6, 8, 10, 12, 14, and 16. Levels were adjusted to a background band marked by an arrowhead. The ORC-induced hypersensitive sites of the post-RC are indicated by asterisks. The functional ARS element is marked by an open box. The position of the ACS is indicated.
Similar articles
- Cdc7 is required throughout the yeast S phase to activate replication origins.
Donaldson AD, Fangman WL, Brewer BJ. Donaldson AD, et al. Genes Dev. 1998 Feb 15;12(4):491-501. doi: 10.1101/gad.12.4.491. Genes Dev. 1998. PMID: 9472018 Free PMC article. - Association of RPA with chromosomal replication origins requires an Mcm protein, and is regulated by Rad53, and cyclin- and Dbf4-dependent kinases.
Tanaka T, Nasmyth K. Tanaka T, et al. EMBO J. 1998 Sep 1;17(17):5182-91. doi: 10.1093/emboj/17.17.5182. EMBO J. 1998. PMID: 9724654 Free PMC article. - Localization of Cdc7 Protein Kinase During DNA Replication in Saccharomyces cerevisiae.
Rossbach D, Bryan DS, Hesselberth JR, Sclafani R. Rossbach D, et al. G3 (Bethesda). 2017 Nov 6;7(11):3757-3774. doi: 10.1534/g3.117.300223. G3 (Bethesda). 2017. PMID: 28924058 Free PMC article. - Functions of mammalian Cdc7 kinase in initiation/monitoring of DNA replication and development.
Kim JM, Yamada M, Masai H. Kim JM, et al. Mutat Res. 2003 Nov 27;532(1-2):29-40. doi: 10.1016/j.mrfmmm.2003.08.008. Mutat Res. 2003. PMID: 14643427 Review. - Cdc7 kinase complex: a key regulator in the initiation of DNA replication.
Masai H, Arai K. Masai H, et al. J Cell Physiol. 2002 Mar;190(3):287-96. doi: 10.1002/jcp.10070. J Cell Physiol. 2002. PMID: 11857444 Review.
Cited by
- Ku complex controls the replication time of DNA in telomere regions.
Cosgrove AJ, Nieduszynski CA, Donaldson AD. Cosgrove AJ, et al. Genes Dev. 2002 Oct 1;16(19):2485-90. doi: 10.1101/gad.231602. Genes Dev. 2002. PMID: 12368259 Free PMC article. - MCM3AP, a novel acetyltransferase that acetylates replication protein MCM3.
Takei Y, Swietlik M, Tanoue A, Tsujimoto G, Kouzarides T, Laskey R. Takei Y, et al. EMBO Rep. 2001 Feb;2(2):119-23. doi: 10.1093/embo-reports/kve026. EMBO Rep. 2001. PMID: 11258703 Free PMC article. - Cell cycle regulation of DNA replication initiator factor Dbf4p.
Cheng L, Collyer T, Hardy CF. Cheng L, et al. Mol Cell Biol. 1999 Jun;19(6):4270-8. doi: 10.1128/MCB.19.6.4270. Mol Cell Biol. 1999. PMID: 10330168 Free PMC article. - Linear derivatives of Saccharomyces cerevisiae chromosome III can be maintained in the absence of autonomously replicating sequence elements.
Dershowitz A, Snyder M, Sbia M, Skurnick JH, Ong LY, Newlon CS. Dershowitz A, et al. Mol Cell Biol. 2007 Jul;27(13):4652-63. doi: 10.1128/MCB.01246-06. Epub 2007 Apr 23. Mol Cell Biol. 2007. PMID: 17452442 Free PMC article. - CDC28 phosphorylates Cac1p and regulates the association of chromatin assembly factor I with chromatin.
Jeffery DC, Kakusho N, You Z, Gharib M, Wyse B, Drury E, Weinreich M, Thibault P, Verreault A, Masai H, Yankulov K. Jeffery DC, et al. Cell Cycle. 2015;14(1):74-85. doi: 10.4161/15384101.2014.973745. Cell Cycle. 2015. PMID: 25602519 Free PMC article.
References
- Amon A, Surana U, Muroff I, Nasmyth K. Regulation of p34CDC28 tyrosine phosphorylation is not required for entry into mitosis in S. cerevisiae. Nature. 1992;355:368–371. - PubMed
- Aparicio OM, Weinstein DM, Bell SP. Components and dynamics of DNA replication complexes in S. cerevisiae: Redistribution of MCM complexes and Cdc45p during S phase. Cell. 1997;91:59–69. - PubMed
- Bahman M, Buck V, White A, Rosamond J. Characterisation of the CDC7 gene product of Saccharomyces cerevisiae as a protein kinase needed for the initiation of mitotic DNA synthesis. Biochim Biophys Acta. 1988;951:335–343. - PubMed
- Bell SP, Kobayashi R, Stillman B. Yeast origin recognition complex functions in transcription silencing and DNA replication. Science. 1993;262:1844–1849. - PubMed
- Brewer BJ, Diller JD, Friedman KL, Kolor KM, Raghuraman MK, Fangman WL. The topography of chromosome replication in yeast. Cold Spring Harbor Symp Quant Biol. 1993;58:425–442. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases