Regulation of riboflavin biosynthesis in Bacillus subtilis is affected by the activity of the flavokinase/flavin adenine dinucleotide synthetase encoded by ribC - PubMed (original) (raw)
Regulation of riboflavin biosynthesis in Bacillus subtilis is affected by the activity of the flavokinase/flavin adenine dinucleotide synthetase encoded by ribC
M Mack et al. J Bacteriol. 1998 Feb.
Abstract
This work shows that the ribC wild-type gene product has both flavokinase and flavin adenine dinucleotide synthetase (FAD-synthetase) activities. RibC plays an essential role in the flavin metabolism of Bacillus subtilis, as growth of a ribC deletion mutant strain was dependent on exogenous supply of FMN and the presence of a heterologous FAD-synthetase gene in its chromosome. Upon cultivation with growth-limiting amounts of FMN, this ribC deletion mutant strain overproduced riboflavin, while with elevated amounts of FMN in the culture medium, no riboflavin overproduction was observed. In a B. subtilis ribC820 mutant strain, the corresponding ribC820 gene product has reduced flavokinase/FAD-synthetase activity. In this strain, riboflavin overproduction was also repressed by exogenous FMN but not by riboflavin. Thus, flavin nucleotides, but not riboflavin, have an effector function for regulation of riboflavin biosynthesis in B. subtilis, and RibC seemingly is not directly involved in the riboflavin regulatory system. The mutation ribC820 leads to deregulation of riboflavin biosynthesis in B. subtilis, most likely by preventing the accumulation of the effector molecule FMN or FAD.
Figures
FIG. 1
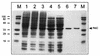
Overproduction and purification of B. subtilis wild-type RibC and mutant RibC820. SDS-PAGE of cell extracts of IPTG-induced E. coli strains harboring the B. subtilis ribC wild-type (E. coli BL21MM01; lane 2) and ribC820 mutant (E. coli BL21MM02; lane 3) genes. As a control, lane 1 shows a cell extract of an IPTG-induced E. coli BL21 strain harboring the expression vector pJF119HE without insert. Lanes 4 and 5 show RibC wild type and mutant, respectively, after (NH4)2SO4 purification. Cation-exchange eluates of RibC wild type and mutant are shown in lanes 6 and 7. Lanes marked M are molecular weight markers (in kilodaltons).
FIG. 2
HPLC chromatograms of the products of flavokinase/FAD-synthetase assays. Assay mixtures containing 50 μM riboflavin, 3 mM ATP, 15 mM MgCl2, and 10 mM sodium sulfite (Na2SO3) were preincubated at 37°C for 5 min. Pure wild-type RibC (A and B) or mutant RibC820 (C and D) (3 μg of each) was added, and the mixtures were incubated for another 5 (A and C) or 30 (B and D) min. An aliquot was removed and separated on an HPLC column (Nucleosil 10 C18; 4.6 by 250 mm; Macherey & Nagel). The following solvent system was used at a flow rate of 2.5 ml/min: 25% (vol/vol) methanol–100 mM formic acid–100 mM ammonium formate (pH 3.7). The reaction was monitored with a fluorescence detector (excitation, 470 nm; emission, 530 nm; Waters Associates). The chromatograms show three clearly resolved peaks of riboflavin (8 min), FMN (5 min), and FAD (4 min). Numbers in parentheses indicate decrease of riboflavin and increase of flavin nucleotides during the enzyme assay.
FIG. 3
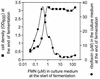
FMN-dependent riboflavin overproduction in B. subtilis 1012MM006. The FMN auxothropic ribC deletion mutant B. subtilis 1012MM006 was cultivated for 16 h in the presence of the indicated amounts of FMN. Riboflavin in the culture medium at the end of fermentation was determined by HPLC analysis as described for Fig. 2. Cell optical density at 600 nm (OD600) at the end of fermentation was estimated photometrically.
FIG. 4
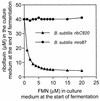
Exogenuous FMN represses riboflavin overproduction in a B. subtilis ribC820 (triangles) but not in a B. subtilis 1012mro87 (ribO mutant, circles) strain. A B. subtilis ribC820 strain and strain 1012mro87 (ribO mutant) were cultivated for 16 h in growth medium supplemented with indicated amounts of FMN. Riboflavin in the supernatant was determined by HPLC as described for Fig. 2. Cell growth was estimated photometrically at 600 nm.
Similar articles
- The bifunctional flavokinase/flavin adenine dinucleotide synthetase from Streptomyces davawensis produces inactive flavin cofactors and is not involved in resistance to the antibiotic roseoflavin.
Grill S, Busenbender S, Pfeiffer M, Köhler U, Mack M. Grill S, et al. J Bacteriol. 2008 Mar;190(5):1546-53. doi: 10.1128/JB.01586-07. Epub 2007 Dec 21. J Bacteriol. 2008. PMID: 18156273 Free PMC article. - The ribR gene encodes a monofunctional riboflavin kinase which is involved in regulation of the Bacillus subtilis riboflavin operon.
Solovieva IM, Kreneva RA, Leak DJ, Perumov DA. Solovieva IM, et al. Microbiology (Reading). 1999 Jan;145 ( Pt 1):67-73. doi: 10.1099/13500872-145-1-67. Microbiology (Reading). 1999. PMID: 10206712 - [Mutational analysis of the ribC gene of Bacillus subtilis].
Karelov DV, Kreneva RA, Érraĭs Lopes L, Perumov DA, Mironov AS. Karelov DV, et al. Genetika. 2011 Jun;47(6):856-61. Genetika. 2011. PMID: 21866869 Russian. - Three biotechnical processes using Ashbya gossypii, Candida famata, or Bacillus subtilis compete with chemical riboflavin production.
Stahmann KP, Revuelta JL, Seulberger H. Stahmann KP, et al. Appl Microbiol Biotechnol. 2000 May;53(5):509-16. doi: 10.1007/s002530051649. Appl Microbiol Biotechnol. 2000. PMID: 10855708 Review. - Enzymes in riboflavin biosynthesis: Potential antibiotic drug targets.
Jaroensuk J, Chuaboon L, Kesornpun C, Chaiyen P. Jaroensuk J, et al. Arch Biochem Biophys. 2023 Oct 15;748:109762. doi: 10.1016/j.abb.2023.109762. Epub 2023 Sep 20. Arch Biochem Biophys. 2023. PMID: 37739114 Review.
Cited by
- Flavoproteins are potential targets for the antibiotic roseoflavin in Escherichia coli.
Langer S, Hashimoto M, Hobl B, Mathes T, Mack M. Langer S, et al. J Bacteriol. 2013 Sep;195(18):4037-45. doi: 10.1128/JB.00646-13. Epub 2013 Jul 8. J Bacteriol. 2013. PMID: 23836860 Free PMC article. - Genomic analysis of a riboflavin-overproducing Ashbya gossypii mutant isolated by disparity mutagenesis.
Kato T, Azegami J, Yokomori A, Dohra H, El Enshasy HA, Park EY. Kato T, et al. BMC Genomics. 2020 Apr 23;21(1):319. doi: 10.1186/s12864-020-6709-7. BMC Genomics. 2020. PMID: 32326906 Free PMC article. - Expression and purification of the 5'-nucleotidase YitU from Bacillus species: its enzymatic properties and possible applications in biotechnology.
Yusupova YR, Skripnikova VS, Kivero AD, Zakataeva NP. Yusupova YR, et al. Appl Microbiol Biotechnol. 2020 Apr;104(7):2957-2972. doi: 10.1007/s00253-020-10428-y. Epub 2020 Feb 10. Appl Microbiol Biotechnol. 2020. PMID: 32040605 Free PMC article. - An _rfuABCD_-Like Operon and Its Relationship to Riboflavin Utilization and Mammalian Infectivity by Borrelia burgdorferi.
Muramatsu MK, Zhou J, Fitzgerald BL, Deka RK, Belisle JT, Norgard MV. Muramatsu MK, et al. Infect Immun. 2021 Sep 16;89(10):e0030721. doi: 10.1128/IAI.00307-21. Epub 2021 Jul 12. Infect Immun. 2021. PMID: 34310888 Free PMC article. - Comprehensive identification of essential Staphylococcus aureus genes using Transposon-Mediated Differential Hybridisation (TMDH).
Chaudhuri RR, Allen AG, Owen PJ, Shalom G, Stone K, Harrison M, Burgis TA, Lockyer M, Garcia-Lara J, Foster SJ, Pleasance SJ, Peters SE, Maskell DJ, Charles IG. Chaudhuri RR, et al. BMC Genomics. 2009 Jul 1;10:291. doi: 10.1186/1471-2164-10-291. BMC Genomics. 2009. PMID: 19570206 Free PMC article.
References
- Arbige M V, Bulthuis B A, Schultz J, Crabb D. Fermentation of Bacillus. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis. Washington, D.C: American Society for Microbiology; 1993. pp. 871–895.
- Bacher A. Biosynthesis of flavins. In: Müller F, editor. Chemistry and biochemistry of flavoenzymes. Vol. 1. Boca Raton, Fla: CRC Press; 1991. pp. 215–259.
- Bacher A. Riboflavin kinase and FAD synthetase. In: Müller F, editor. Chemistry and biochemistry of flavoenzymes. Vol. 1. Boca Raton, Fla: CRC Press; 1991. pp. 349–370.
- Bacher A, Eberhardt S, Richter G. Biosynthesis of riboflavin. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: ASM Press; 1996. pp. 657–664.
- Bacher A, Eisenreich K, Kis K, Ladenstein R, Richter G, Scheuring J, Weinkauf S. Biosynthesis of flavins. In: Dugas H, Schmidtchen F P, editors. Bioorganic chemistry frontiers. Vol. 3. Berlin, Germany: Springer-Verlag; 1993. pp. 147–192.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases