beta-D-glucosyl-hydroxymethyluracil is a conserved DNA modification in kinetoplastid protozoans and is abundant in their telomeres - PubMed (original) (raw)
beta-D-glucosyl-hydroxymethyluracil is a conserved DNA modification in kinetoplastid protozoans and is abundant in their telomeres
F van Leeuwen et al. Proc Natl Acad Sci U S A. 1998.
Abstract
The unusual DNA base beta-D-glucosyl-hydroxymethyluracil, called "J, " replaces approximately 0.5-1% of Thy in DNA of African trypanosomes but has not been found in other organisms thus far. In Trypanosoma brucei, J is located predominantly in repetitive DNA, and its presence correlates with the silencing of telomeric genes. Using antibodies specific for J, we have developed sensitive assays to screen for J in a range of organisms and have found that J is not limited to trypanosomes that undergo antigenic variation but is conserved among Kinetoplastida. In all kinetoplastids tested, including the human pathogens Leishmania donovani and Trypanosoma cruzi, J was found to be abundantly present in the (GGGTTA)n telomere repeats. Outside Kinetoplastida, J was found only in Diplonema, a small phagotrophic marine flagellate, in which we also identified 5-MeCyt. Fractionation of Diplonema DNA showed that the two modifications are present in a common genome compartment, which suggests that they may have a similar function. Dinoflagellates appear to contain small amounts of modified bases that may be analogs of J. The evolutionary conservation of J in kinetoplastid protozoans suggests that it has a general function, repression of transcription or recombination, or a combination of both. T. brucei may have recruited J for the control of genes involved in antigenic variation.
Figures
Figure 1
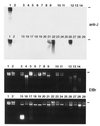
Conservation of J in telomeric repeats of Kinetoplastida. (A) Dot blot with 200 ng of DNA of each kinetoplastid sample indicated was incubated with anti-J antiserum (anti-J). Abbreviations of the organisms and the life cycle stages are explained in Table 1. Bound antibodies were detected with a second antibody conjugated to horseradish peroxidase and were visualized by enhanced chemiluminescence. After stripping the blot, DNA loading was checked by hybridization by using a (GGGTTA)5-telomeric repeat oligo as a common probe (telomeric repeats). J also was found in L. tarentolae, L. donovani chagasi, L. brasiliensis, L. mexicana, and T. vivax (data not shown). (B) An anti-J immunoprecipitation of sonicated DNA. Modified DNA bound by antibody (ip) and 10% of the supernatant (10% sup) were blotted and analyzed by hybridization with the telomeric repeat probe.
Figure 2
Screening for J in DNA with a zoo blot. Southern blot of a 1% agarose gel with ≈200 ng of total DNA of each sample was incubated with antiserum 539αJ, and bound antibodies were indirectly detected by enhanced chemiluminescence (anti-J). Black lines indicate the position of the slots. Lanes: 1, bloodstream form T. brucei; 2, procyclic (insect form) T. brucei; 3–6, human blood, sperm, breast tumor, and ovarian tumor; 7–8, mouse testis and lung; 9, calf thymus; 10–11, human cell lines HeLa and G401; 12–14, human DNAs enriched ≈100-fold for telomeric repeats; 12, telomeric tracts from HeLa cells;13, matrix-attached DNA from HeLa cells;14, telomeric tracts from G401 cells;15, D. melanogaster (also contained RNA); 16, Sf9 cells; 17, C. elegans; 18–19, S. cerevisiae strains M398 and BJ1991; 20, P. pastoris; 21, P. micans; 22, C. cohnii; 23, P. falciparum; 24, T. gondii; 25, E. histolytica; 26, Diplonema; 27, T. vaginalis; 28, G. lamblia; and 29, E. coli. Because no common probe was available, DNA loading was checked by the staining of the gel with ethidium bromide (EtBr).
Figure 3
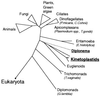
Phylogenetic tree of eukaryotes tested for the presence of J. This phylogenetic tree inferred from 16S-like rRNA sequence similarities is modified from Sogin (39). The position of Diplonema (dashed line) is still unclear (see text). J was found in the order Kinetoplastida and in Diplonema (underlined).
Figure 4
Detection of J in Diplonema and J-like modifications in dinoflagellates. Analysis of bloodstream form T. brucei, P. micans, C. cohnii, and Diplonema by 32P-nucleotide postlabeling combined with 2D-TLC (D1 and D2 are indicated). The position of the labeled 5′-deoxynucleotidemonophosphates (dN) is explained in the right bottom corner. a, c, u, and g indicate contaminating ribonucleotides. dU and HOMedU nucleotides comigrate under these conditions. (Top) Labeling of total DNA. J is indicated with small arrows. (Middle) Analysis of samples after anti-J immunoprecipitation of sonicated DNA fragments. Arrowheads indicate nucleotides in dinoflagellates that migrate close to, but differently from, J. To test whether the nucleotide close to dC in Diplonema is 5-Me-dCMP, it was isolated together with dCMP and CMP, chemically deaminated (+), and rerun on 2D-TLC, mixed with nondeaminated input (−). Deamination of 5-Me-dCMP, dCMP, and CMP results in dTMP, dUMP, and UMP, respectively. (Bottom) Labeled nucleotides from the Middle were incubated with anti-J antibodies coupled to ProtA beads to specifically remove nucleotides recognized by the antibodies. The supernatant was analyzed by 2D-TLC.
Comment in
- A base called J.
Simpson L. Simpson L. Proc Natl Acad Sci U S A. 1998 Mar 3;95(5):2037-8. doi: 10.1073/pnas.95.5.2037. Proc Natl Acad Sci U S A. 1998. PMID: 9482833 Free PMC article. Review. No abstract available.
Similar articles
- beta-D-glucosyl-hydroxymethyluracil, a novel base in African trypanosomes and other Kinetoplastida.
Borst P, van Leeuwen F. Borst P, et al. Mol Biochem Parasitol. 1997 Dec 1;90(1):1-8. doi: 10.1016/s0166-6851(97)00170-9. Mol Biochem Parasitol. 1997. PMID: 9497027 Review. - The telomeric GGGTTA repeats of Trypanosoma brucei contain the hypermodified base J in both strands.
van Leeuwen F, Wijsman ER, Kuyl-Yeheskiely E, van der Marel GA, van Boom JH, Borst P. van Leeuwen F, et al. Nucleic Acids Res. 1996 Jul 1;24(13):2476-82. doi: 10.1093/nar/24.13.2476. Nucleic Acids Res. 1996. PMID: 8692684 Free PMC article. - Base J originally found in kinetoplastida is also a minor constituent of nuclear DNA of Euglena gracilis.
Dooijes D, Chaves I, Kieft R, Dirks-Mulder A, Martin W, Borst P. Dooijes D, et al. Nucleic Acids Res. 2000 Aug 15;28(16):3017-21. doi: 10.1093/nar/28.16.3017. Nucleic Acids Res. 2000. PMID: 10931915 Free PMC article. - Tandemly repeated DNA is a target for the partial replacement of thymine by beta-D-glucosyl-hydroxymethyluracil in Trypanosoma brucei.
van Leeuwen F, Kieft R, Cross M, Borst P. van Leeuwen F, et al. Mol Biochem Parasitol. 2000 Jul;109(2):133-45. doi: 10.1016/s0166-6851(00)00247-4. Mol Biochem Parasitol. 2000. PMID: 10960172 - A base called J.
Simpson L. Simpson L. Proc Natl Acad Sci U S A. 1998 Mar 3;95(5):2037-8. doi: 10.1073/pnas.95.5.2037. Proc Natl Acad Sci U S A. 1998. PMID: 9482833 Free PMC article. Review. No abstract available.
Cited by
- Glucosylated hydroxymethyluracil, DNA base J, prevents transcriptional readthrough in Leishmania.
van Luenen HG, Farris C, Jan S, Genest PA, Tripathi P, Velds A, Kerkhoven RM, Nieuwland M, Haydock A, Ramasamy G, Vainio S, Heidebrecht T, Perrakis A, Pagie L, van Steensel B, Myler PJ, Borst P. van Luenen HG, et al. Cell. 2012 Aug 31;150(5):909-21. doi: 10.1016/j.cell.2012.07.030. Cell. 2012. PMID: 22939620 Free PMC article. - Epigenetic Regulation of Transcription in Trypanosomatid Protozoa.
Martínez-Calvillo S, Romero-Meza G, Vizuet-de-Rueda JC, Florencio-Martínez LE, Manning-Cela R, Nepomuceno-Mejía T. Martínez-Calvillo S, et al. Curr Genomics. 2018 Feb;19(2):140-149. doi: 10.2174/1389202918666170911163517. Curr Genomics. 2018. PMID: 29491742 Free PMC article. Review. - Defining the sequence requirements for the positioning of base J in DNA using SMRT sequencing.
Genest PA, Baugh L, Taipale A, Zhao W, Jan S, van Luenen HG, Korlach J, Clark T, Luong K, Boitano M, Turner S, Myler PJ, Borst P. Genest PA, et al. Nucleic Acids Res. 2015 Feb 27;43(4):2102-15. doi: 10.1093/nar/gkv095. Epub 2015 Feb 6. Nucleic Acids Res. 2015. PMID: 25662217 Free PMC article. - Facile enzymatic synthesis of base J-containing oligodeoxyribonucleotides and an analysis of the impact of base J on DNA replication in cells.
Ji D, Wang Y. Ji D, et al. PLoS One. 2014 Jul 25;9(7):e103335. doi: 10.1371/journal.pone.0103335. eCollection 2014. PLoS One. 2014. PMID: 25061973 Free PMC article. - Regulation of transcription termination by glucosylated hydroxymethyluracil, base J, in Leishmania major and Trypanosoma brucei.
Reynolds D, Cliffe L, Förstner KU, Hon CC, Siegel TN, Sabatini R. Reynolds D, et al. Nucleic Acids Res. 2014 Sep;42(15):9717-29. doi: 10.1093/nar/gku714. Epub 2014 Aug 7. Nucleic Acids Res. 2014. PMID: 25104019 Free PMC article.
References
- Gommers-Ampt J H, van Leeuwen F, de Beer A L, Vliegenthart J F, Dizdaroglu M, Kowalak J A, Crain P F, Borst P. Cell. 1993;75:1129–1136. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases