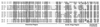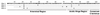Evolutionary analyses of the 12-kDa acidic ribosomal P-proteins reveal a distinct protein of higher plant ribosomes - PubMed (original) (raw)
Evolutionary analyses of the 12-kDa acidic ribosomal P-proteins reveal a distinct protein of higher plant ribosomes
K Szick et al. Proc Natl Acad Sci U S A. 1998.
Abstract
The P-protein complex of eukaryotic ribosomes forms a lateral stalk structure in the active site of the large ribosomal subunit and is thought to assist in the elongation phase of translation by stimulating GTPase activity of elongation factor-2 and removal of deacylated tRNA. The complex in animals, fungi, and protozoans is composed of the acidic phosphoproteins P0 (35 kDa), P1 (11-12 kDa), and P2 (11-12 kDa). Previously we demonstrated by protein purification and microsequencing that ribosomes of maize (Zea mays L.) contain P0, one type of P1, two types of P2, and a distinct P1/P2 type protein designated P3. Here we implemented distance matrices, maximum parsimony, and neighbor-joining analyses to assess the evolutionary relationships between the 12 kDa P-proteins of maize and representative eukaryotic species. The analyses identify P3, found to date only in mono- and dicotyledonous plants, as an evolutionarily distinct P-protein. Plants possess three distinct groups of 12 kDa P-proteins (P1, P2, and P3), whereas animals, fungi, and protozoans possess only two distinct groups (P1 and P2). These findings demonstrate that the P-protein complex has evolved into a highly divergent complex with respect to protein composition despite its critical position within the active site of the ribosome.
Figures
Figure 1
Alignment of a subset of archaebacterial and eukaryotic 12 kDa P-proteins. The amino acid sequences represent archaebacteria [Sulfolobus solfataricus (Sso), Halobacterium marismortui (Hma)], yeast [Saccharomyces cerevisiae (Sce)], rat (Rattus rattus (Rra)], and plants [Zea mays (Zma), Oryza sativa (Osa), Arabidopsis thaliana (Ath)]. Sequences were aligned using
clustal w
(49) and were adjusted manually upon visual inspection. Gaps were introduced to ensure maximum homology. Amino acids of conserved physicochemical similarity are shaded based on the following criteria: (i) conserved amino acids must occur in three of the four P-protein groups (i.e., L12, P1, P2, and P3) and (ii) conserved amino acids must occur in at least 75% of the sequences. The N termini, acidic hinge regions, and highly conserved C termini of the 12-kDa P-proteins are indicated.
Figure 2
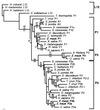
Amino acid phylogenetic analysis of the 12 kDa P-proteins. A phylogenetic tree was generated by the neighbor-joining method using Kimura-corrected distances in
phylip
version 3.5c based on the amino acid sequence of 33 eukaryotic 12 kDa P-proteins and 4 archaebacterial L12 proteins. Branch lengths are proportional to the amino acid distances along each branch. Bootstrap values from 500 replicates are indicated. Parsimony bootstrap values for clades supported above the 50% level are indicated below branches, whereas neighbor-joining bootstrap values based on Kimura-corrected distances or uncorrected distances are indicated above the branch (corrected distances are in boldface italics).
Figure 3
Maize genomic DNA Southern blot analysis. Genomic DNA digested with _Bam_HI (B), _Eco_RI (E), or _Hin_dIII (H) was separated, blotted, and hybridized at high stringency with the [32P]-labeled maize cDNAs encoding P1, P2a, P2b, and P3, respectively. (A) rpp1, (B) rpp2a-1, (C) rpp2b, (D) rpp3. Migration of DNA standards (in kilobases) are indicated.
Figure 4
Maize P2a deduced peptide sequence. Deduced peptide sequences of the maize P2a family members were aligned using the
pileup
alignment program (28). Amino acids that differ from P2a-1 are indicated. Gaps were introduced to ensure maximum homology.
Figure 5
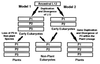
Alternative models depicting the evolution of the plant P3 protein. Model 1. Duplication and divergence of the ancestral L12-like gene occurred very early in the eukaryotic lineage resulting in P1, P2, and P3 type proteins in ancestral eukaryotes. Contemporary plants have retained the P3 gene, whereas the specific loss of P3 from other contemporary eukaryotes is necessary to explain the absence of P3 in these species. Model 2. Duplication and divergence of the ancestral L12-like gene occurred very early in the eukaryotic lineage to produce P1 and P2 type P-proteins in ancestral eukaryotes. Further duplication and divergence within the plant lineage produced the P3 seen in modern plants.
Similar articles
- Acidic phosphoprotein complex of the 60S ribosomal subunit of maize seedling roots. Components and changes in response to flooding.
Bailey-Serres J, Vangala S, Szick K, Lee CH. Bailey-Serres J, et al. Plant Physiol. 1997 Aug;114(4):1293-305. doi: 10.1104/pp.114.4.1293. Plant Physiol. 1997. PMID: 9276949 Free PMC article. - In vivo formation of Plasmodium falciparum ribosomal stalk - a unique mode of assembly without stable heterodimeric intermediates.
Wawiórka L, Krokowski D, Gordiyenko Y, Krowarsch D, Robinson CV, Adam I, Grankowski N, Tchórzewski M. Wawiórka L, et al. Biochim Biophys Acta. 2015 Jan;1850(1):150-8. doi: 10.1016/j.bbagen.2014.10.015. Epub 2014 Oct 23. Biochim Biophys Acta. 2015. PMID: 25450178 - A mode of assembly of P0, P1, and P2 proteins at the GTPase-associated center in animal ribosome: in vitro analyses with P0 truncation mutants.
Hagiya A, Naganuma T, Maki Y, Ohta J, Tohkairin Y, Shimizu T, Nomura T, Hachimori A, Uchiumi T. Hagiya A, et al. J Biol Chem. 2005 Nov 25;280(47):39193-9. doi: 10.1074/jbc.M506050200. Epub 2005 Sep 27. J Biol Chem. 2005. PMID: 16188884 - Proteins P1, P2, and P0, components of the eukaryotic ribosome stalk. New structural and functional aspects.
Remacha M, Jimenez-Diaz A, Santos C, Briones E, Zambrano R, Rodriguez Gabriel MA, Guarinos E, Ballesta JP. Remacha M, et al. Biochem Cell Biol. 1995 Nov-Dec;73(11-12):959-68. doi: 10.1139/o95-103. Biochem Cell Biol. 1995. PMID: 8722011 Review. - Structure and function of the acidic ribosomal stalk proteins.
Wahl MC, Möller W. Wahl MC, et al. Curr Protein Pept Sci. 2002 Feb;3(1):93-106. doi: 10.2174/1389203023380756. Curr Protein Pept Sci. 2002. PMID: 12370014 Review.
Cited by
- The organization of cytoplasmic ribosomal protein genes in the Arabidopsis genome.
Barakat A, Szick-Miranda K, Chang IF, Guyot R, Blanc G, Cooke R, Delseny M, Bailey-Serres J. Barakat A, et al. Plant Physiol. 2001 Oct;127(2):398-415. Plant Physiol. 2001. PMID: 11598216 Free PMC article. - Systematic Review of Plant Ribosome Heterogeneity and Specialization.
Martinez-Seidel F, Beine-Golovchuk O, Hsieh YC, Kopka J. Martinez-Seidel F, et al. Front Plant Sci. 2020 Jun 25;11:948. doi: 10.3389/fpls.2020.00948. eCollection 2020. Front Plant Sci. 2020. PMID: 32670337 Free PMC article. - The Arabidopsis Cytosolic Ribosomal Proteome: From form to Function.
Carroll AJ. Carroll AJ. Front Plant Sci. 2013 Mar 1;4:32. doi: 10.3389/fpls.2013.00032. eCollection 2013. Front Plant Sci. 2013. PMID: 23459595 Free PMC article. - Erythrocytic stage-dependent regulation of oligomerization of Plasmodium ribosomal protein P2.
Das S, Sudarsan R, Sivakami S, Sharma S. Das S, et al. J Biol Chem. 2012 Nov 30;287(49):41499-513. doi: 10.1074/jbc.M112.384388. Epub 2012 Oct 11. J Biol Chem. 2012. PMID: 23060439 Free PMC article. - Extraribosomal Functions of Cytosolic Ribosomal Proteins in Plants.
Xiong W, Lan T, Mo B. Xiong W, et al. Front Plant Sci. 2021 Apr 21;12:607157. doi: 10.3389/fpls.2021.607157. eCollection 2021. Front Plant Sci. 2021. PMID: 33968093 Free PMC article. Review.
References
- Nygård O, Nilsson L. Eur J Biochem. 1990;191:1–17. - PubMed
- Hershey J W B. Annu Rev Biochem. 1991;69:717–755. - PubMed
- Merrick W C, Hershey J W B. Translational Control. Plainview, NY: Cold Spring Harbor Lab. Press; 1995.
- Wool I G, Chan Y-L, Gluck A. Biochem Cell Biol. 1995;73:933–947. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources