Identification of a substrate-targeting domain in cyclin E necessary for phosphorylation of the retinoblastoma protein - PubMed (original) (raw)
Identification of a substrate-targeting domain in cyclin E necessary for phosphorylation of the retinoblastoma protein
B L Kelly et al. Proc Natl Acad Sci U S A. 1998.
Abstract
Considerable advances have been made in characterizing the cyclins and cyclin-dependent kinases (CDKs) that are necessary for progression through the cell cycle, but there has been relatively lesser success in identifying the specific biochemical pathways and cell cycle events that are directly under CDK control. To identify physiologically significant CDK substrates we generated mutations in cyclin E that altered the ability of the cyclin to direct the cyclin-CDK holoenzyme to specific in vivo substrates. We show that one of these mutations defines a domain in cyclin E necessary for phosphorylation of the retinoblastoma protein (Rb). These observations confirm the idea that cyclins contribute to substrate recognition by cyclin-CDK complexes, demonstrate the utility of targeting mutants in the identification of essential cyclin-CDK substrates, and put cyclin E squarely into the family of proteins designed to regulate Rb.
Figures
Figure 1
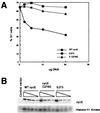
Transfection of wild-type and mutant cyclin E proteins into NIH 3T3 cells. NIH 3T3 cells were transiently transfected with increasing amounts of myc-tagged wild-type or mutant cyclin E. (A) Percentage of cells in G1 phase of the cell cycle versus amount of transfected plasmid. (B) Cyclin E protein expression and associated histone H1 kinase activity from cells depicted in A.
Figure 2
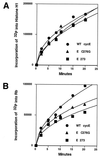
Transfection of wild-type and mutant cyclin E proteins into SAOS-2 cells. (A) Percentage of cells in G1 phase of the cell cycle versus amount of transfected plasmid. (B) SAOS-2 cells transfected with pCMVpRb plasmid and increasing amounts of cyclin E: percentage of cells in G1 phase of the cell cycle versus amount of transfected plasmid. (C) Cyclin E-associated histone H1 kinase activity from cells transfected with pCMVpRb plus cyclin E (D) Phosphorylation of transfected full-length Rb by cyclin E (5 μg of transfected DNA) in SAOS-2 cells.
Figure 3
Mutant cyclin E proteins cannot overcome a p16 cell cycle block in NIH 3T3 cells. (A) NIH 3T3 cells were transfected with cyclin E in the presence or absence of p16. For each transfected population, DNA content is shown on the x axis and cell number on the y axis. (B) Transient transfection of NIH 3T3 cells comparing in vivo phosphorylation of exogenous large pocket Rb by cyclin E in the presence of exogenous p16.
Figure 4
VxCxE mutants are defective for Rb but not histone H1 phosphorylation in vitro. (A) Recombinant cyclin E–CDK2 complexes made in baculovirus-infected Sf-9 insect cells were incubated with [γ-32P]ATP and either histone H1 (A) or Rb (B) for increasing amounts of time. Results shown are representative of three different experiments.
Figure 5
Binding of cyclin E to Rb in vivo and in vitro. (A) Cyclin E associated with either wild-type or catalytically inactive CDK2 was mixed with recombinant Rb in the presence and absence of ATP, as indicated. Stable binding of Rb to cyclin E was determined by coimmunoprecipitation using anti-cyclin E antibodies. (B) Lysates of Sf-9 cells infected with inactive CDK2 and either wild-type or ΔVDCLE cyclin E were mixed with recombinant Rb. Binding of Rb to cyclin E was determined by coimmunoprecipitation using anti-cyclin Rb antibodies. (C) Wild-type cyclin E, Rb, and the indicated CDK2 proteins were expressed in NIH 3T3 cells and 293 cells. Binding of Rb to cyclin E was determined as in B. (D) Wild-type or mutant cyclin E, Rb, and the indicated CDK2 protein were expressed in 293 cells. Binding of Rb to cyclin E was determined as in B. In this experiment binding of Rb to cyclin E in the absence of CDK2 is not as evident as in C.
References
- Sherr C J. Cell. 1994;79:551–555. - PubMed
- Geng Y, Eaton E N, Picon M, Roberts J M, Lundberg A S, Gifford A, Sardet C, Weinberg R A. Oncogene. 1996;12:1173–1180. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases