Mas oncogene signaling and transformation require the small GTP-binding protein Rac - PubMed (original) (raw)
Mas oncogene signaling and transformation require the small GTP-binding protein Rac
I E Zohn et al. Mol Cell Biol. 1998 Mar.
Abstract
The Mas oncogene encodes a novel G-protein-coupled receptor that was identified originally as a transforming protein when overexpressed in NIH 3T3 cells. The mechanism and signaling pathways that mediate Mas transformation have not been determined. We observed that the foci of transformed NIH 3T3 cells caused by Mas were similar to those caused by activated Rho and Rac proteins. Therefore, we determined if Mas signaling and transformation are mediated through activation of a specific Rho family protein. First, we observed that, like activated Rac1, Mas cooperated with activated Raf and caused synergistic transformation of NIH 3T3 cells. Second, both Mas- and Rac1-transformed NIH 3T3 cells retained actin stress fibers and showed enhanced membrane ruffling. Third, like Rac, Mas induced lamellipodium formation in porcine aortic endothelial cells. Fourth, Mas and Rac1 strongly activated the JNK and p38, but not ERK, mitogen-activated protein kinases. Fifth, Mas and Rac1 stimulated transcription from common DNA promoter elements: NF-kappaB, serum response factor (SRF), Jun/ATF-2, and the cyclin D1 promoter. Finally, Mas transformation and some of Mas signaling (SRF and cyclin D1 but not NF-kappaB activation) were blocked by dominant negative Rac1. Taken together, these observations suggest that Mas transformation is mediated in part by activation of Rac-dependent signaling pathways. Thus, Rho family proteins are common mediators of transformation by a diverse variety of oncogene proteins that include Ras, Dbl family, and G-protein-coupled oncogene proteins.
Figures
FIG. 1
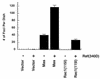
Mas-transformed cells exhibit a transformed phenotype similar to those of Rac- and Rho-transformed cells. (A) Transformed foci from NIH 3T3 cultures transfected with pZIP-ras(61L), pZIP-rhoA(63L), and pZIP-mas. (B) Morphology of NIH 3T3 cells stably transfected with pZIP-NeoSV(x)1, pZIP-ras(61L), pZIP-rac1(115I), and pZIP-mas. Multiple (>100) G418-resistant colonies were pooled to establish the cell lines used for these analyses.
FIG. 2
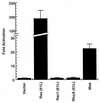
Like Rac1, Mas cooperates with Raf(340D) and causes synergistic focus-forming activity. NIH 3T3 cells were cotransfected with pZIP expression plasmids encoding the indicated proteins. One hundred nanograms of pZIP-mas cDNA and 1 μg of all other DNAs were transfected per 60-mm-diameter dish. The data are shown as mean ± standard error for triplicate plates and are representative of at least three separate experiments.
FIG. 3
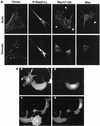
Mas and Rac1 cause similar changes in the organization of the actin cytoskeleton. (A) Mas- and Rac-transformed cells retain stress fibers and show enhanced membrane ruffling. Stably transfected NIH 3T3 cell lines expressing the indicated proteins were subject to immunofluorescence analysis as described in Materials and Methods. Shown are actin stress fibers and focal adhesions stained with RITC-phalloidin and FITC-antivinculin antibodies, respectively. Membrane ruffles are indicated by arrowheads. (B) Like Rac1(12V), Mas induces membrane ruffles in PAE cells. PAE cells were microinjected with expression constructs encoding either Mas along with GFP (a and b) or Myc epitope-tagged Rac1(12V) (c and d) as described in Materials and Methods. Cells were serum starved following injection, and actin was stained with RITC-phalloidin (a and c). Microinjected cells were identified by expression of GFP (for Mas) (b) or anti-Myc antiserum (for Rac1) (d).
FIG. 4
Mas and Rac are strong activators of p38 and JNK but not ERK. (A) Activation of ERK2 by Mas. Cos-7 cells were transfected with either pCGN-hyg (vector), pCGN-ras(61L), pCGN-_rac_1(61L), or pCGN-mas along with an HA epitope-tagged ERK2 expression vector. Immunocomplex kinase assays with myelin basic protein (MBP) as a substrate were performed following immunoprecipitation of HA-ERK2 (top panel). Fold activation (Act) (middle panel) of ERK was determined by PhosphorImager analysis and expressed relative to phosphorylation levels in vector-transfected cells. Twenty-five micrograms of lysate was resolved by SDS-PAGE, transferred to an Immobilon membrane, and subsequently probed with anti-HA antibody to ensure equivalent expression levels of HA-ERK2 (bottom panel). (B) Activation of JNK1 by Mas. Cos-7 cells were transfected as for panel A but with FLAG epitope-tagged JNK1. JNK1 kinase activity was determined with GST–c-Jun(1-79) as a substrate (top panel). Fold activation (middle panel) and JNK1 levels (bottom panel) were determined as described for panel A. Data in panels A and B are representative of at least three independent experiments in Cos-7 and NIH 3T3 cells, using pCGN and pCDNA3 expression plasmids. (C) Activation of p38 by Mas. Cos-7 cells were transfected as described for panels A and B but with FLAG epitope-tagged p38. p38 kinase activity was determined with GST–ATF-2(1-254) as a substrate (top panel); fold activation (middle panel) and p38 expression levels (bottom panel) were determined as described above. Data are representative of two separate experiments.
FIG. 5
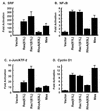
Mas and Rac stimulate transcription from common promoter elements. NIH 3T3 cells were transfected with pAX142 (vector), pAX142-ras(61L), pAX142-rac1(61L), pAX142-rhoA(63L), or pAX142-mas along with luciferase gene reporter constructs for SRF transcriptional activity (A), NF-κB transcriptional activity (B), c-Jun/ATF-2 transcriptional activity (C), and cyclin D1 expression (D). Data shown are representative of at least three independent experiments using both pAX142 and pCDNA3 mammalian expression constructs.
FIG. 6
Mas, but not Rac1, caused activation of Elk-1. NIH 3T3 cells were transfected with pAX142 (vector), pAX142-ras(61L), pAX142-rac1(61L), pAX142-rhoA(63L), or pAX142-mas along with Gal4–Elk-1 and the Gal4-responsive 5XGal4-Luc construct. Data shown are representative of at least three independent experiments using both pAX142 and pCDNA3 mammalian expression constructs.
FIG. 7
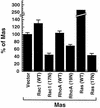
Dominant negative Rac1 blocks Mas signaling. NIH 3T3 cells were cotransfected with pAX142-mas and either pAX142-rac1(17N) or pAX142-rac1(WT) along with luciferase gene reporter constructs for SRF (A), cyclin D1 (B), or NF-κB (C) expression. Data are expressed as the mean of the percentage of the activation in the Mas-plus-vector samples ± standard deviation of duplicate samples and are representative of at least two independent experiments.
FIG. 8
Dominant negative Rac, RhoA, and Ras block Mas transformation. NIH 3T3 cells were transfected with pZIP-mas and the indicated wild-type (WT) and dominant negative Rho family proteins, and the focus formation assay was performed as described in Materials and Methods. Data are expressed as the mean of the percentage of the total number of foci in the Mas-plus-vector dishes ± standard error and are the average of six separate experiments performed in duplicate or triplicate.
Similar articles
- Rac regulation of transformation, gene expression, and actin organization by multiple, PAK-independent pathways.
Westwick JK, Lambert QT, Clark GJ, Symons M, Van Aelst L, Pestell RG, Der CJ. Westwick JK, et al. Mol Cell Biol. 1997 Mar;17(3):1324-35. doi: 10.1128/MCB.17.3.1324. Mol Cell Biol. 1997. PMID: 9032259 Free PMC article. - Rit, a non-lipid-modified Ras-related protein, transforms NIH3T3 cells without activating the ERK, JNK, p38 MAPK or PI3K/Akt pathways.
Rusyn EV, Reynolds ER, Shao H, Grana TM, Chan TO, Andres DA, Cox AD. Rusyn EV, et al. Oncogene. 2000 Sep 28;19(41):4685-94. doi: 10.1038/sj.onc.1203836. Oncogene. 2000. PMID: 11032018 - The small GTPases Cdc42Hs, Rac1 and RhoG delineate Raf-independent pathways that cooperate to transform NIH3T3 cells.
Roux P, Gauthier-Rouvière C, Doucet-Brutin S, Fort P. Roux P, et al. Curr Biol. 1997 Sep 1;7(9):629-37. doi: 10.1016/s0960-9822(06)00289-2. Curr Biol. 1997. PMID: 9285711 - Rho family proteins and Ras transformation: the RHOad less traveled gets congested.
Zohn IM, Campbell SL, Khosravi-Far R, Rossman KL, Der CJ. Zohn IM, et al. Oncogene. 1998 Sep 17;17(11 Reviews):1415-38. doi: 10.1038/sj.onc.1202181. Oncogene. 1998. PMID: 9779988 Review. - Regulation of phosphorylation pathways by p21 GTPases. The p21 Ras-related Rho subfamily and its role in phosphorylation signalling pathways.
Lim L, Manser E, Leung T, Hall C. Lim L, et al. Eur J Biochem. 1996 Dec 1;242(2):171-85. doi: 10.1111/j.1432-1033.1996.0171r.x. Eur J Biochem. 1996. PMID: 8973630 Review.
Cited by
- Anti-cancer potential of persimmon (Diospyros kaki) leaves via the PDGFR-Rac-JNK pathway.
Kim HS, Suh JS, Jang YK, Ahn SH, Raja G, Kim JC, Jung Y, Jung SH, Kim TJ. Kim HS, et al. Sci Rep. 2020 Oct 22;10(1):18119. doi: 10.1038/s41598-020-75140-3. Sci Rep. 2020. PMID: 33093618 Free PMC article. - International Union of Basic and Clinical Pharmacology. XCIX. Angiotensin Receptors: Interpreters of Pathophysiological Angiotensinergic Stimuli [corrected].
Karnik SS, Unal H, Kemp JR, Tirupula KC, Eguchi S, Vanderheyden PM, Thomas WG. Karnik SS, et al. Pharmacol Rev. 2015 Oct;67(4):754-819. doi: 10.1124/pr.114.010454. Pharmacol Rev. 2015. PMID: 26315714 Free PMC article. Review. - Renin-angiotensin system in the kidney: What is new?
Ferrão FM, Lara LS, Lowe J. Ferrão FM, et al. World J Nephrol. 2014 Aug 6;3(3):64-76. doi: 10.5527/wjn.v3.i3.64. World J Nephrol. 2014. PMID: 25332897 Free PMC article. Review. - Angiotensin-(1-7) is an endogenous ligand for the G protein-coupled receptor Mas.
Santos RA, Simoes e Silva AC, Maric C, Silva DM, Machado RP, de Buhr I, Heringer-Walther S, Pinheiro SV, Lopes MT, Bader M, Mendes EP, Lemos VS, Campagnole-Santos MJ, Schultheiss HP, Speth R, Walther T. Santos RA, et al. Proc Natl Acad Sci U S A. 2003 Jul 8;100(14):8258-63. doi: 10.1073/pnas.1432869100. Epub 2003 Jun 26. Proc Natl Acad Sci U S A. 2003. PMID: 12829792 Free PMC article. - 2020 update on the renin-angiotensin-aldosterone system in pediatric kidney disease and its interactions with coronavirus.
Simões E Silva AC, Lanza K, Palmeira VA, Costa LB, Flynn JT. Simões E Silva AC, et al. Pediatr Nephrol. 2021 Jun;36(6):1407-1426. doi: 10.1007/s00467-020-04759-1. Epub 2020 Sep 29. Pediatr Nephrol. 2021. PMID: 32995920 Free PMC article. Review.
References
- Albanese C, Johnson J, Watanabe G, Eklund N, Vu D, Arnold A, Pestell R G. Transforming p21ras mutants and c-Ets-2 activate the cyclin D1 promoter through distinguishable regions. J Biol Chem. 1995;270:23589–23597. - PubMed
- Andrawis N S, Dzau V J, Pratt R E. Autocrine stimulation of mas oncogene leads to altered growth control. Cell Biol Int Rep. 1992;16:547–556. - PubMed
- Angel P, Hattori K, Smeal T, Karin M. The jun proto-oncogene is positively autoregulated by its product, Jun/AP-1. Cell. 1988;55:875–885. - PubMed
- Baichwal V R, Baeuerle P A. Apoptosis: activate NF-KB or die? Curr Biol. 1997;7:R94–R96. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA042978/CA/NCI NIH HHS/United States
- CA63071/CA/NCI NIH HHS/United States
- CA55008/CA/NCI NIH HHS/United States
- CA42978/CA/NCI NIH HHS/United States
- R01 CA063071/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous