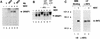Interferon regulatory factor 3 and CREB-binding protein/p300 are subunits of double-stranded RNA-activated transcription factor DRAF1 - PubMed (original) (raw)
Interferon regulatory factor 3 and CREB-binding protein/p300 are subunits of double-stranded RNA-activated transcription factor DRAF1
B K Weaver et al. Mol Cell Biol. 1998 Mar.
Abstract
Cells respond to viral infection or double-stranded RNA with the transcriptional induction of a subset of alpha/beta interferon-stimulated genes by a pathway distinct from the interferon signal pathway. The transcriptional induction is mediated through a DNA sequence containing the alpha/beta interferon-stimulated response element (ISRE). We previously identified a novel transcription factor, designated double-stranded RNA-activated factor 1 (DRAF1), that recognizes this response element. The DNA-binding specificity of DRAF1 correlates with transcriptional induction, thereby distinguishing it as a positive regulator of alpha/beta interferon-stimulated genes. Two of the components of DRAF1 have now been identified as interferon regulatory factor 3 (IRF-3) and the transcriptional coactivator CREB-binding protein (CBP)/p300. We demonstrate that IRF-3 preexists in the cytoplasm of uninfected cells and translocates to the nucleus following viral infection. Translocation of IRF-3 is accompanied by an increase in serine and threonine phosphorylation. Coimmunoprecipitation analyses of endogenous proteins demonstrate an association of IRF-3 with the transcriptional coactivators CBP and p300 only subsequent to infection. In addition, antibodies to the IRF-3, CBP, and p300 molecules react with DRAF1 bound to the ISRE target site of induced genes. The cellular response that leads to DRAF1 activation and specific gene expression may serve to increase host survival during viral infection.
Figures
FIG. 1
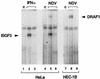
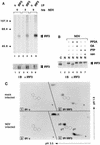
Activation of ISRE-binding factors by IFN-α and NDV. HeLa cells were either untreated (control [c]; lanes 1 and 4), treated with IFN-α for 1 h (lanes 2 and 3), or infected with NDV for 3 h (lanes 5 and 6). HEC-1B cells were either untreated (lane 7) or infected with NDV for 3 h (lanes 8 and 9). Nuclear extracts were prepared and analyzed by an EMSA with a radiolabeled ISRE probe. The specificity of the protein-DNA complexes was shown by competition with a 100-fold excess of unlabeled ISRE oligonucleotide (lanes 3, 6, and 9).
FIG. 2
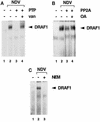
(A) Effect of a protein tyrosine phosphatase (PTP) on DRAF1. HEC-1B nuclear extracts from untreated cells (lane 1) or cells infected with NDV (lanes 2 to 4) were used in an EMSA with a radiolabeled ISRE probe. Prior to the DNA-binding reaction, the extracts were incubated in the absence (lanes 1 and 2) or in the presence (lanes 3 and 4) of recombinant PTP1B. Tyrosine phosphatase activity was inhibited by the inclusion of 5 mM sodium vanadate (van) (lane 4). (B) Effect of a serine/threonine protein phosphatase on DRAF1. HEC-1B nuclear extracts from untreated cells (lane 1) or cells infected with NDV (lanes 2 to 4) were used in an EMSA. Prior to the DNA-binding reaction, the extracts were incubated in the absence (lanes 1 and 2) or in the presence (lanes 3 and 4) of PP2A. Protein phosphatase activity was inhibited by the inclusion of 10 nM okadaic acid (OA) (lane 4). (C) Effect of protein alkylation on the appearance of DRAF1. HEC-1B nuclear extracts from untreated cells (lane 1) or cells infected with NDV (lanes 2 and 3) were used in an EMSA. Prior to the DNA-binding reaction, the extracts were either untreated (lanes 1 and 2) or treated with 10 mM NEM (lane 3).
FIG. 3
(A) Effect of antibody to IRF-3 on DRAF1. Nuclear extracts from untreated HEC-1B cells (lanes 1 to 3) or cells infected with NDV (lanes 4 to 6) were used in an EMSA with a radiolabeled ISRE probe. Prior to the addition of the probe, either no antisera (Ab) (lanes 1 and 4), control murine antisera (c) (1:50 dilution) (lanes 2 and 5), or specific murine antisera to IRF-3 (1:50 dilution) (lanes 3 and 6) were added to the DNA-binding reaction mixtures. (B) Inhibition by GST–IRF-3 fusion protein of anti–IRF-3 antibody effects. Nuclear extracts from untreated HEC-1B cells (lane 1) or cells infected with NDV (lanes 2 to 7) were used in an EMSA. Prior to the addition of the probe, either 1 μg of control rabbit antibody (Ab) (lane 3) or 1 μg of rabbit antibody to IRF-3 (lanes 4 to 7) was added. Antibody incubation was done in the absence of added GST fusion protein (lanes 3 and 4), in the presence of 200 ng (lane 5) and 600 ng (lane 6) of GST–IRF-3 (amino acids 107 to 208), or in the presence of 200 ng of GST-p48 (amino acids 103 to 225) (lane 7). (C) Effect of NDV infection on the ability of IRF-3 to bind to the ISRE. Whole-cell extracts were prepared from HEC-1B cells that were either untreated (lanes 1 and 3) or infected with NDV for 6 h (lanes 2 and 4). The extracts were incubated with ISRE-Sepharose beads (lanes 1 and 2) or with rabbit antibody to IRF-3 (lanes 3 and 4). The bound proteins were separated by SDS-PAGE and detected by immunoblotting (IB) with a murine antibody (α) to IRF-3. IP, immunoprecipitation. Positions of prestained molecular mass standards are shown in kilodaltons on the left.
FIG. 4
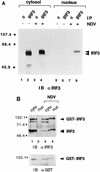
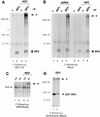
(A) Cellular localization of IRF-3 in control and virus-infected cells. Nuclear and cytoplasmic extracts were prepared from untreated HEC-1B cells (lanes 1, 2, 5, and 6) or cells infected with NDV for 6 h (lanes 3, 4, 7, and 8). Extracts were normalized for cell equivalents and subjected to immunoprecipitation (IP) with control rabbit antibody (c) (lanes 1, 3, 5, and 7) or rabbit antibody to IRF-3 (lanes 2, 4, 6, and 8). The immunoprecipitated proteins were separated by SDS-PAGE and detected by immunoblotting (IB) with a murine antibody (α) to IRF-3. Numbers at left are in kilodaltons. (B) Cellular localization of a transfected GST–IRF-3 chimera. Nuclear (nuc) and cytoplasmic (cyto) extracts were prepared from untreated HT1080/GST–IRF-3 cells (lanes 1 and 2) or cells infected with NDV for 6 h (lanes 3 and 4). Extracts were normalized for cell equivalents and subjected to immunoprecipitation with a rabbit antibody to IRF-3. The immunoprecipitated proteins were separated by SDS-PAGE and detected by immunoblotting (IB) with a murine antibody (α) to IRF-3 (upper panel). The blot was subsequently stripped and reprobed with a rabbit antibody (α) to GST (lower panel).
FIG. 5
(A) Increase in IRF-3 phosphorylation during viral infection. Whole-cell extracts were prepared by detergent lysis of HEC-1B cells that were metabolically labeled with [32P]orthophosphate for a total of 7 h during either mock infection (0 h) (lanes 1 and 2) or infection with NDV for 3 h (lanes 3 and 4) or 6 h (lanes 5 and 6). Extracts were subjected to immunoprecipitation (IP) with a control murine antibody (c) (lanes 1, 3, and 5) or a murine anti–IRF-3 antibody (lanes 2, 4, and 6). Immunoprecipitated proteins were denatured in SDS sample buffer, and 75% of the sample was separated by SDS-PAGE. The gel was fixed, dried, and exposed to film (upper panel). The remaining 25% was analyzed by SDS-PAGE and immunoblotting (IB) with a rabbit polyclonal antibody (α) to IRF-3 (lower panel). The positions of the prestained molecular mass standards are shown in kilodaltons on the left. (B) Effect of protein phosphatase treatment on the migration of IRF-3 in SDS-PAGE. Cytoplasmic (C) and nuclear (N) extracts were prepared from HEC-1B cells that were either untreated (lanes 1 and 2) or infected with NDV (lanes 3 to 7). A total of 100 μg of cytoplasmic extract or 35 μg of nuclear extract was incubated in the absence of phosphatase (lanes 1 to 3), in the presence of PP2A (lanes 4 and 5), or in the presence of PTP1B (PTP) (lanes 6 and 7). The activities of PP2A and PTP1B were inhibited by the inclusion of 10 nM okadaic acid (OA) (lane 5) and 5 mM sodium vanadate (van) (lane 7), respectively. Proteins were separated by SDS-PAGE and detected by immunoblotting (IB) with a murine antibody (α) to IRF-3. (C) Phosphoamino acid analysis of 32P-labeled IRF-3. Cells were metabolically labeled with [32P]orthophosphate, and lysates from mock-infected cells (upper panels) or NDV-infected cells (lower panels) were immunoprecipitated (IP) with control (c) antibody (boxes 1 and 3) or antibody to IRF-3 (boxes 2 and 4). Following SDS-PAGE, proteins were electroblotted to Immobilon-P membranes, and the area containing IRF-3 or the adjacent area from control immunoprecipitations was subjected to partial acid hydrolysis. The hydrolysates were resolved by two-dimensional electrophoresis on a TLC plate. Positions of the serine, threonine, and tyrosine phosphoamino acid internal standards are indicated by dotted circles (pS, pT, and pY, respectively), and origins are denoted by a plus sign.
FIG. 6
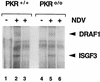
Activation of DRAF1 in _Pkr_0/0 MEFs. Wild-type (Pkr+/+) MEFs or PKR-deficient (_Pkr_0/0) MEFs were either untreated (lanes 1 and 4) or infected with NDV for 3 h (lanes 2, 3, 5, and 6). Nuclear extracts were prepared and analyzed by an EMSA with a radiolabeled ISRE probe. The specificity of the protein-DNA complexes was shown by competition with a 100-fold excess of unlabeled ISRE oligonucleotide (lanes 3 and 6).
FIG. 7
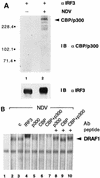
(A) Association of IRF-3 with a 300-kDa cellular protein following HEC-1B cell infection with NDV. Detergent cell lysates were prepared from cells following metabolic labeling with [35S]methionine-[35S]cysteine during either mock infection (lanes 1 and 2) or infection with NDV for 6 h (lanes 3 and 4). Extracts were subjected to immunoprecipitation (IP) with a control rabbit antibody (c) (lanes 1 and 3) or a rabbit anti–IRF-3 antibody (lanes 2 and 4). The immunoprecipitated proteins were separated by SDS-PAGE and detected by fluorography. The positions of the prestained molecular mass standards are shown in kilodaltons on the left. (B) Presence in HeLa cells treated with dsRNA or infected with NDV of a 300-kDa cellular protein associated with IRF-3. Detergent cell lysates were prepared from cells that were metabolically labeled with [35S]methionine-[35S]cysteine. Cells were untreated (lanes 1 and 2), treated with 150 μg of poly(rI) · poly(rC) per ml for 2 h (lanes 3 and 4), mock infected (lanes 5 and 6), or infected with NDV for 6 h (lanes 7 and 8). The immunoprecipitation analysis was performed as described in panel A. (C) Association of the IRF-3-associated 300-kDa protein with the ISRE following virus infection. Whole-cell extracts were prepared from HEC-1B cells that were metabolically labeled with [35S]methionine-[35S]cysteine and either mock infected (lanes 1 and 2) or infected with NDV for 6 h (lanes 3 and 4). The extracts were incubated with DNA-Sepharose beads prepared with native ISRE (wt) (lanes 2 and 4) or with a mutated ISRE (mt) (lanes 1 and 3). The DNA-bound proteins were detected as described in panel A. (D) Copurification of the 300-kDa protein with stably expressed GST–IRF-3 fusion protein. Detergent cell lysates were prepared from HT1080/GST–IRF-3 cells that were metabolically labeled with [35S]methionine-[35S]cysteine and either mock infected (lane 1) or infected with NDV for 6 h (lane 2). Extracts were incubated with glutathione-agarose beads and washed, and bound proteins were separated and detected as described in panel A. Asterisks indicate migration of 300-kDa proteins.
FIG. 8
(A) Identification of the IRF-3-associated protein as CBP/p300. Detergent cell lysates were prepared from HEC-1B cells that were either mock infected (lane 1) or infected with NDV for 6 h (lane 2). Extracts were subjected to immunoprecipitation with rabbit anti–IRF-3 antibody (α). Immunoprecipitated proteins were separated by SDS-PAGE and detected by immunoblotting (IB) with a mixture of specific rabbit antibodies to p300 and CBP (upper panel) or with a murine antibody to IRF-3 (lower panel). Numbers at left are in kilodaltons. (B) Presence of CBP and p300 in the DRAF1 complex. Nuclear extracts from untreated HEC-1B cells (lane 1) or cells infected with NDV (lanes 2 to 10) were used in an EMSA with a radiolabeled ISRE probe. Prior to the addition of the probe, no antibody (Ab) (lanes 1 and 2), 2 μg of control rabbit antibody (c) (lane 3), 2 μg of specific rabbit antibody to IRF-3 (lane 4), 2 μg of specific rabbit antibody to p300 (lanes 5 and 8), 2 μg of specific rabbit antibody to CBP (lanes 6 and 9), or 1 μg of antibodies to both p300 and CBP (lanes 7 and 10) was added to the DNA-binding reaction mixtures. The effect of the anti-p300 and anti-CBP antibodies was blocked by the inclusion of 0.2 μg of control peptide(s) used as an immunogen to generate the antibodies (lanes 8 to 10).
Similar articles
- Regulated nuclear-cytoplasmic localization of interferon regulatory factor 3, a subunit of double-stranded RNA-activated factor 1.
Kumar KP, McBride KM, Weaver BK, Dingwall C, Reich NC. Kumar KP, et al. Mol Cell Biol. 2000 Jun;20(11):4159-68. doi: 10.1128/MCB.20.11.4159-4168.2000. Mol Cell Biol. 2000. PMID: 10805757 Free PMC article. - Apoptosis is promoted by the dsRNA-activated factor (DRAF1) during viral infection independent of the action of interferon or p53.
Weaver BK, Ando O, Kumar KP, Reich NC. Weaver BK, et al. FASEB J. 2001 Feb;15(2):501-15. doi: 10.1096/fj.00-0222com. FASEB J. 2001. PMID: 11156966 - Analyses of virus-induced homomeric and heteromeric protein associations between IRF-3 and coactivator CBP/p300.
Suhara W, Yoneyama M, Iwamura T, Yoshimura S, Tamura K, Namiki H, Aimoto S, Fujita T. Suhara W, et al. J Biochem. 2000 Aug;128(2):301-7. doi: 10.1093/oxfordjournals.jbchem.a022753. J Biochem. 2000. PMID: 10920266 - Control of IRF-3 activation by phosphorylation.
Yoneyama M, Suhara W, Fujita T. Yoneyama M, et al. J Interferon Cytokine Res. 2002 Jan;22(1):73-6. doi: 10.1089/107999002753452674. J Interferon Cytokine Res. 2002. PMID: 11846977 Review. - Triggering the interferon response: the role of IRF-3 transcription factor.
Hiscott J, Pitha P, Genin P, Nguyen H, Heylbroeck C, Mamane Y, Algarte M, Lin R. Hiscott J, et al. J Interferon Cytokine Res. 1999 Jan;19(1):1-13. doi: 10.1089/107999099314360. J Interferon Cytokine Res. 1999. PMID: 10048763 Review.
Cited by
- ISG15/USP18/STAT2 is a molecular hub regulating IFN I-mediated control of Dengue and Zika virus replication.
Espada CE, da Rocha EL, Ricciardi-Jorge T, Dos Santos AA, Soares ZG, Malaquias G, Patrício DO, Gonzalez Kozlova E, Dos Santos PF, Bordignon J, Sanford TJ, Fajardo T, Sweeney TR, Báfica A, Mansur DS. Espada CE, et al. Front Immunol. 2024 Feb 7;15:1331731. doi: 10.3389/fimmu.2024.1331731. eCollection 2024. Front Immunol. 2024. PMID: 38384473 Free PMC article. - Innate cellular response to virus particle entry requires IRF3 but not virus replication.
Collins SE, Noyce RS, Mossman KL. Collins SE, et al. J Virol. 2004 Feb;78(4):1706-17. doi: 10.1128/jvi.78.4.1706-1717.2004. J Virol. 2004. PMID: 14747536 Free PMC article. - Regulation of DNA-raised immune responses by cotransfected interferon regulatory factors.
Sasaki S, Amara RR, Yeow WS, Pitha PM, Robinson HL. Sasaki S, et al. J Virol. 2002 Jul;76(13):6652-9. doi: 10.1128/jvi.76.13.6652-6659.2002. J Virol. 2002. PMID: 12050378 Free PMC article. - Antiviral actions of interferons.
Samuel CE. Samuel CE. Clin Microbiol Rev. 2001 Oct;14(4):778-809, table of contents. doi: 10.1128/CMR.14.4.778-809.2001. Clin Microbiol Rev. 2001. PMID: 11585785 Free PMC article. Review. - The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3.
Basler CF, Mikulasova A, Martinez-Sobrido L, Paragas J, Mühlberger E, Bray M, Klenk HD, Palese P, García-Sastre A. Basler CF, et al. J Virol. 2003 Jul;77(14):7945-56. doi: 10.1128/jvi.77.14.7945-7956.2003. J Virol. 2003. PMID: 12829834 Free PMC article.
References
- Bandyopadhyay S K, Leonard G T, Jr, Bandyopadhyay T, Stark G R, Sen G C. Transcriptional induction by double-stranded RNA is mediated by interferon-stimulated response elements without activation of interferon-stimulated gene factor 3. J Biol Chem. 1995;270:19624–19629. - PubMed
- Bannister A J, Kourarides T. The CBP coactivator is a histone acetyltransferase. Nature. 1996;384:641–643. - PubMed
- Bhattacharya S, Eckner R, Grossman S, Oldread E, Arany Z, D’Andrea A, Livingston D M. Cooperation of Stat2 and p300/CBP in signalling induced by interferon-α. Nature. 1996;383:344–347. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous