Signal peptidase cleavage at the flavivirus C-prM junction: dependence on the viral NS2B-3 protease for efficient processing requires determinants in C, the signal peptide, and prM - PubMed (original) (raw)
Signal peptidase cleavage at the flavivirus C-prM junction: dependence on the viral NS2B-3 protease for efficient processing requires determinants in C, the signal peptide, and prM
C E Stocks et al. J Virol. 1998 Mar.
Abstract
Signal peptidase cleavage at the C-prM junction in the flavivirus structural polyprotein is inefficient in the absence of the cytoplasmic viral protease, which catalyzes cleavage at the COOH terminus of the C protein. The signal peptidase cleavage occurs efficiently in circumstances where the C protein is deleted or if the viral protease complex is present. In this study, we used cDNA of Murray Valley encephalitis virus (MVE) to examine features of the structural polyprotein which allow this regulation of a luminal cleavage by a cytoplasmic protease. We found that the inefficiency of signal peptidase cleavage in the absence of the viral protease is not attributable solely to features of the C protein. Inhibition of cleavage still occurred when charged residues in C were mutated to uncharged residues or when an unrelated protein sequence (that of ubiquitin) was substituted for C. Also, fusion of the C protein did not inhibit processing of an alternative adjacent signal sequence. The cleavage region of the flavivirus prM translocation signal is unusually hydrophobic, and we established that altering this characteristic by making three point mutations near the signal peptidase cleavage site in MVE prM dramatically increased the extent of cleavage without requiring removal of the C protein. In addition, we demonstrated that luminal sequences downstream from the signal peptidase cleavage site contributed to the inefficiency of cleavage.
Figures
FIG. 1
Schematic diagram showing the locations of alterations to charged residues in derivatives of the MVE C protein. Plasmid pSTR encodes the C-prM-E-NS1-NS2A-6%NS2B region of the MVE polyprotein. The C protein is represented by a solid line, and the beginning of the prM translocation signal is indicated by a dashed line. Positions of charged residues are indicated by + for Arg or Lys and − for Asp or Glu. Residues 1 to 23 and 97 to 105 of the C protein encoded by this construct are shown in single-letter amino acid code. Residues in these regions which are unchanged in the charge mutant derivatives of pSTR are denoted by dots. Sequences outside the marked regions were identical in all constructs. The site of cleavage by the viral NS2B-3 protease is marked with an arrow.
FIG. 2
Substitution of basic residues in the C protein does not influence signal peptidase cleavage at the C-prM junction. (A) COS cells were transfected with pSTR (lanes 1 and 5), pSTR.Cmut2aa (lanes 2 and 6), pSTR.Cmut4aa (lanes 3 and 7), or pSTR.Cmut11aa (lanes 4 and 8), with (+) or without (−) pNS3/T. Lane 9 shows the results of transfection with pNS3/T only. Two days after transfection, the monolayers were metabolically labelled for 30 min and cell lysates were subjected to immunoprecipitation with anti-MVE mouse ascitic fluid. Polypeptides were separated by SDS-PAGE (12% polyacrylamide). Bands corresponding to MVE E, NS1, prM, and putative C-prM glycoproteins are labelled, and the numbers indicate the sizes (in kilodaltons) of marker proteins in lane M. (B) Schematic diagrams showing the expected topology at the ER membrane of polyproteins encoded by pSTR variants in the presence and absence of NS2B-3. M denotes the ER membrane, L denotes the ER lumen, and Cy denotes the cytoplasmic side of the membrane. Sites of efficient cleavage by signal peptidase are indicated by arrowheads, the proteolytic cleavage catalyzed by the viral NS2B-3 complex is denoted by an arrow, and regions of the MVE C protein which were altered in the various pSTR.Cmut constructs are indicated by circles in parentheses.
FIG. 3
Replacement of the C protein with ubiquitin at the NH2 terminus of the prM signal sequence does not alter the efficiency of prM cleavage by signal peptidase. (A) COS cells were transiently transfected with the ubiquitin fusion construct pSTR.UbmutV (lanes 1 and 4) or pSTR.Ub (lanes 2 and 5), in which the cleavage site for ubiquitin-specific proteases was destroyed or left intact, respectively, or with pNS3/T (lanes 3 and 6). The cells were metabolically labelled for 30 min, and polypeptides immunoprecipitated from lysates with anti-MVE mouse ascitic fluid (lanes 1 to 3) or antiserum against a ubiquitin–glutathione _S_-transferase fusion protein (lanes 4 to 6) were separated by SDS-PAGE (12% polyacrylamide) (lanes 1 to 3) or SDS-PAGE (15% polyacrylamide) (lanes 4 to 6). Bands corresponding to MVE E, NS1, and prM glycoproteins, as well as ubiquitin (Ub) and putative Ub-prM fusion products, are labelled, and the numbers indicate the sizes (in kilodaltons) of marker proteins in lane M. (B) Schematic diagrams showing the expected topology at the ER membrane of polyproteins encoded by pSTR.UbmutV and pSTR.Ub. Abbreviations are as listed in the legend to Fig. 2. Ubiquitin-specific protease cleavage is indicated by a dotted arrow, and a solid circle shows the approximate position of the Gly76→Val substitution mutation.
FIG. 4
NH2-terminal linkage of the MVE C protein to an idealized signal sequence does not inhibit cleavage by signal peptidase. (A) COS cells transfected with pSFVΔE3.MVE-C (lane 1), pSFVΔE3 (lane 3), or pNS3/T (lane 4) or cotransfected with pSFVΔE3.MVE-C and pNS3/T (lane 2) were metabolically labelled for 3 h. The products immunoprecipitated by an anti-SFV mouse serum were separated by SDS-PAGE (10% polyacrylamide) under nonreducing conditions to separate E2 and E1. Bands corresponding to the SFV E2 and E1 glycoproteins are shown, and the numbers indicate the positions and sizes (in kilodaltons) of marker proteins. (B) Schematic diagrams showing the expected topology at the ER membrane of polyproteins encoded by pSFVΔE3.MVE-C and pSFVΔE3. Abbreviations are as listed in the legend to Fig. 2. Autocatalytic cleavage of the unmutated SFV C protein is indicated by a curved arrow.
FIG. 5
Alignment of flavivirus amino acid sequences around the C-prM junction. The MVE prM translocation signal with flanking sequences encoded by pSTR is shown at the top, with the sequences we have considered to be the h- and c-regions indicated. Corresponding sequences are also shown for yellow fever virus (YF) (32), Japanese encephalitis virus (JE) (38), Kunjin virus (KUN) (7), tick-borne encephalitis virus (TBE) (26), dengue virus type 2 (DEN2) (13) and West Nile virus (WN) (6). Large arrows indicate sites of cleavage by the viral NS2B-3 protease complex and signal peptidase. Small arrows show sites at which cleavage potential scores based on the algorithm of von Heijne (39) are greater than an arbitrary value of 4. Positively (+) and negatively (−) charged amino acid residues are labelled.
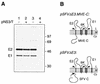
FIG. 6
Mutations in the signal sequence of prM induce efficient signal peptidase cleavage independent of the presence of the NS2B-3 protease. (A) COS cells transiently transfected with pSTR (lane 1), pSTR.mutPQAQA (lane 2), or pSTR.ΔC (lane 3) were metabolically labelled for 3 h. Polypeptides immunoprecipitated from cell lysates with anti-MVE mouse ascitic fluid were resolved by SDS-PAGE (12% polyacrylamide). Bands corresponding to MVE E, NS1, and prM glycoproteins are labelled, and the numbers indicate the positions and sizes (in kilodaltons) of marker proteins. (B) Schematic diagrams showing the expected topology at the ER membrane of polyproteins encoded by pSTR.mutPQAQA and pSTRΔC. Abbreviations are as listed in the legend to Fig. 2. The location of the altered amino acid residues encoded by pSTR.mutPQAQA is represented by a solid circle.
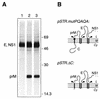
FIG. 7
The production of MVE E from a Canch-E fusion protein is dependent on the viral protease. (A) COS cells transiently transfected with pCanch-E (lanes 1 and 2) or pSTR (lanes 3 and 4), in the absence (−) or presence (+) of pNS3/T, were metabolically labelled for 30 min. Products immunoprecipitated from the lysates with anti-MVE E monoclonal antibody M2-8E7 (14) were examined by SDS-PAGE (12% polyacrylamide). Positions of E, prM (coprecipitated as part of a prM-E heterodimer), and putative Canch-E fusion glycoproteins are labelled, and the numbers indicate the positions and sizes (in kilodaltons) of marker proteins. (B) Schematic diagram showing the expected topology at the ER membrane of the polyprotein encoded by pCanch-E. Abbreviations are as listed in the legend to Fig. 2.
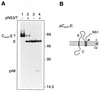
FIG. 8
The production of H-2Kd from a Canch-Kd fusion protein is not dependent on the viral protease. (A) COS cells were transiently transfected with pKd (lane 1), pCanch-Kd (lanes 2 and 3), or pCanch-KdmutK (lanes 4 and 5), with (+) or without (−) pNS3/T. Lane 6 shows the results of transfection with pNS3/T only. Cell monolayers were pulse-labelled for 30 min, and the products immunoprecipitated from cell lysates with an anti-Kd monoclonal antibody (Hb159) were subjected to SDS-PAGE (12% polyacrylamide). Bands corresponding to the Kd and possible Canch-Kd fusion proteins are labelled, and the numbers indicate the positions and sizes (in kilodaltons) of marker proteins. (B) Schematic diagrams showing the expected topology at the ER membrane of polyproteins encoded by pKd, pCanch-Kd and pCanch-KdmutK. Abbreviations are as listed in the legend to Fig. 2, and the Pro→Lys substitution encoded in pCanch-KdmutK is represented by a solid circle.
Similar articles
- Formation of the flavivirus envelope: role of the viral NS2B-NS3 protease.
Yamshchikov VF, Compans RW. Yamshchikov VF, et al. J Virol. 1995 Apr;69(4):1995-2003. doi: 10.1128/JVI.69.4.1995-2003.1995. J Virol. 1995. PMID: 7884844 Free PMC article. - NS2B/3 proteolysis at the C-prM junction of the tick-borne encephalitis virus polyprotein is highly membrane dependent.
Kurz M, Stefan N, Zhu J, Skern T. Kurz M, et al. Virus Res. 2012 Sep;168(1-2):48-55. doi: 10.1016/j.virusres.2012.06.012. Epub 2012 Jun 19. Virus Res. 2012. PMID: 22727684 Free PMC article. - Mutagenesis of the signal sequence of yellow fever virus prM protein: enhancement of signalase cleavage In vitro is lethal for virus production.
Lee E, Stocks CE, Amberg SM, Rice CM, Lobigs M. Lee E, et al. J Virol. 2000 Jan;74(1):24-32. doi: 10.1128/jvi.74.1.24-32.2000. J Virol. 2000. PMID: 10590087 Free PMC article. - Delayed by Design: Role of Suboptimal Signal Peptidase Processing of Viral Structural Protein Precursors in Flaviviridae Virus Assembly.
Alzahrani N, Wu MJ, Shanmugam S, Yi M. Alzahrani N, et al. Viruses. 2020 Sep 26;12(10):1090. doi: 10.3390/v12101090. Viruses. 2020. PMID: 32993149 Free PMC article. Review. - Post-translational regulation and modifications of flavivirus structural proteins.
Roby JA, Setoh YX, Hall RA, Khromykh AA. Roby JA, et al. J Gen Virol. 2015 Jul;96(Pt 7):1551-69. doi: 10.1099/vir.0.000097. Epub 2015 Feb 23. J Gen Virol. 2015. PMID: 25711963 Review.
Cited by
- Pan-serotype dengue virus inhibitor JNJ-A07 targets NS4A-2K-NS4B interaction with NS2B/NS3 and blocks replication organelle formation.
Kiemel D, Kroell AH, Denolly S, Haselmann U, Bonfanti JF, Andres JI, Ghosh B, Geluykens P, Kaptein SJF, Wilken L, Scaturro P, Neyts J, Van Loock M, Goethals O, Bartenschlager R. Kiemel D, et al. Nat Commun. 2024 Jul 19;15(1):6080. doi: 10.1038/s41467-024-50437-3. Nat Commun. 2024. PMID: 39030239 Free PMC article. - Interaction between hTIM-1 and Envelope Protein Is Important for JEV Infection.
Liang Z, Pan J, Xie S, Yang X, Cao R. Liang Z, et al. Viruses. 2023 Jul 21;15(7):1589. doi: 10.3390/v15071589. Viruses. 2023. PMID: 37515282 Free PMC article. - Development of a novel virus-like particle-based vaccine for preventing tick-borne encephalitis virus infection.
Tang J, Fu M, Xu C, Xue B, Zhou A, Chen S, Zhao H, Zhou Y, Chen J, Yang Q, Chen X. Tang J, et al. Virol Sin. 2023 Oct;38(5):767-777. doi: 10.1016/j.virs.2023.06.003. Epub 2023 Jun 14. Virol Sin. 2023. PMID: 37328107 Free PMC article. - An optimized messenger RNA vaccine candidate protects non-human primates from Zika virus infection.
Bollman B, Nunna N, Bahl K, Hsiao CJ, Bennett H, Butler S, Foreman B, Burgomaster KE, Aleshnick M, Kong WP, Fisher BE, Ruckwardt TJ, Morabito KM, Graham BS, Dowd KA, Pierson TC, Carfi A. Bollman B, et al. NPJ Vaccines. 2023 Apr 20;8(1):58. doi: 10.1038/s41541-023-00656-4. NPJ Vaccines. 2023. PMID: 37080988 Free PMC article. - Assembly-defective Tembusu virus ectopically expressing capsid protein is an approach for live-attenuated flavivirus vaccine development.
He Y, Guo J, Wang X, Zhang S, Mao L, Hu T, Wang M, Jia R, Zhu D, Liu M, Zhao X, Yang Q, Wu Y, Zhang S, Huang J, Mao S, Ou X, Gao Q, Sun D, Cheng A, Chen S. He Y, et al. NPJ Vaccines. 2022 May 12;7(1):51. doi: 10.1038/s41541-022-00468-y. NPJ Vaccines. 2022. PMID: 35550523 Free PMC article.
References
- Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. - PubMed
- Baker R T, Smith S A, Marano R, McKee J, Board P G. Protein expression using cotranslational fusion and cleavage of ubiquitin. J Biol Chem. 1994;269:25381–25386. - PubMed
- Brodsky J L. Post-translational protein translocation: not all hsc70s are created equal. Trends Biochem Sci. 1996;21:122–126. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources