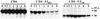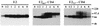A retention signal necessary and sufficient for endoplasmic reticulum localization maps to the transmembrane domain of hepatitis C virus glycoprotein E2 - PubMed (original) (raw)
A retention signal necessary and sufficient for endoplasmic reticulum localization maps to the transmembrane domain of hepatitis C virus glycoprotein E2
L Cocquerel et al. J Virol. 1998 Mar.
Abstract
The hepatitis C virus (HCV) genome encodes two envelope glycoproteins (E1 and E2). These glycoproteins interact to formin a noncovalent heterodimeric complex which is retained in the endoplasmic reticulum (ER). To identify whether E1 and/or E2 contains an ER-targeting signal potentially involved in ER retention of the E1-E2 complex, these proteins were expressed alone and their intracellular localization was studied. Due to misfolding of E1 in the absence of E2, no conclusion on the localization of its native form could be drawn from the expression of E1 alone. E2 expressed in the absence of E1 was shown to be retained in the ER similarly to E1-E2 complex. Chimeric proteins in which E2 domains were exchanged with corresponding domains of a protein normally transported to the plasma membrane (CD4) were constructed to identify the sequence responsible for its ER retention. The transmembrane domain (TMD) of E2 (C-terminal 29 amino acids) was shown to be sufficient for retention of the ectodomain of CD4 in the ER compartment. Replacement of the E2 TMD by the anchor signal of CD4 or a glycosyl phosphatidylinositol (GPI) moiety led to its expression on the cell surface. In addition, replacement of the E2 TMD by the anchor signal of CD4 or a GPI moiety abolished the formation of E1-E2 complexes. Together, these results suggest that, besides having a role as a membrane anchor, the TMD of E2 is involved in both complex formation and intracellular localization.
Figures
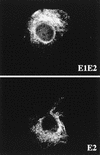
FIG. 1
Localization by indirect immunofluorescence of E2, expressed in the presence or absence of E1. CV-1 cells were coinfected with vTF7-3 and either vHCV170-809 (E1-E2) or vHCV371-809 (E2) at a multiplicity of infection of 1 PFU/cell. Cells were fixed with isopropanol at 12 h postinfection and immunostained with anti-E2 MAb H53.
FIG. 2
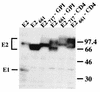
Sensitivity of E2 to endo H treatment. HepG2 cells were coinfected with vTF7-3 and vaccinia virus recombinants expressing either E1-E2 (vHCV170-809), E2 (vHCV371-809), or a truncated form of E2 (vHCV371-661) at a multiplicity of infection of 5 PFU/cell. Infected cells were pulse-labeled for 10 min and chased for the indicated times (in hours). Cell lysates were immunoprecipitated with MAb H53 and then treated or not with endo H. Samples were separated by SDS-PAGE (10% polyacrylamide). Deglycosylated proteins are indicated by asterisks. The sizes (in kilodaltons) of protein molecular mass markers are indicated on the right. The predicted sequence of E2 contains 11 N-linked potential glycosylation sites.
FIG. 3
Schematic representation of the parental proteins E2 and CD4 and chimeric proteins used in this study drawn to scale. E2661-CD4, ectodomain of E2, lacking its C-terminal 56 amino acids, fused to the TMD and cytoplasmic domain of CD4; E2717-CD4, ectodomain of E2 fused to the TMD and cytoplasmic domain of CD4; CD4-E2662, ectodomain of CD4 fused to the C-terminal 85 amino acids of E2; CD4-E2718, ectodomain of CD4 fused to the TMD of E2 (C-terminal 29 amino acids); E2661-GPI, ectodomain of E2, lacking its C-terminal 56 amino acids, fused to the C-terminal 37 amino acids of DAF; E2717-GPI, ectodomain of E2 fused to the C-terminal 37 amino acids of DAF. The ectodomain of E2 is from amino acid 384 to 717 (position on the polyprotein) and the TMD of E2 from amino acid 718 to 746. The ectodomain of CD4 is from amino acid 1 to 373, its TMD is from amino acid 374 to 395, and its cytosolic domain is from amino acid 396 to 435. Details on constructions are reported in Materials and Methods. (B) C-terminal portions of E2661-GPI and E2717-GPI. The amino acid sequences identify the junction between the E2 ectodomain or its truncated form and the 37-amino-acid GPI anchor addition sequence from DAF. Between these two sequences, there is a sequence of two additional amino acids resulting from cloning. The asterisk below the Ser in the DAF sequence indicates the predicted point of attachment for the GPI anchor (34).
FIG. 4
Expression of CD4, CD4-E2662, and CD4-E2718 analyzed in pulse-chase experiments. HepG2 cells were coinfected with vTF7-3 and the appropriate vaccinia virus recombinant at a multiplicity of infection of 5 PFU/cell. Infected cells were pulse-labeled for 10 min and chased for the indicated times (in hours). Cell lysates were immunoprecipitated with MAb OKT4 (anti-CD4). Samples were separated by SDS-PAGE (10% polyacrylamide). The sizes (in kilodaltons) of protein molecular mass markers are indicated on the right.
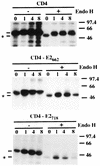
FIG. 5
Sensitivities of CD4-E2662 and CD4-E2718 to endo H treatment. HepG2 cells were coinfected with vTF7-3 and the appropriate vaccinia virus recombinant at a multiplicity of infection of 5 PFU/cell. Infected cells were pulse-labeled for 10 min and chased for the indicated times (in hours). Cell lysates were immunoprecipitated with MAb OKT4 and then treated or not with endo H. Samples were separated by SDS-PAGE (10% polyacrylamide). Deglycosylated proteins are indicated by asterisks. The sizes (in kilodaltons) of protein molecular mass markers are indicated on the right.
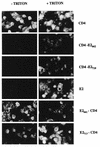
FIG. 6
Cell surface expression of chimeric proteins analyzed by indirect immunofluorescence. HepG2 cells were coinfected with vTF7-3 and the appropriate vaccinia virus recombinant at a multiplicity of infection of 1 PFU/cell. Cells were fixed with paraformaldehyde at 12 h postinfection, permeabilized or not with Triton X-100, and immunostained with anti-E2 (E2, E2661-CD4, and E2717-CD4) or anti-CD4 (CD4, CD4-E2662, and CD4-E2718) antibodies.
FIG. 7
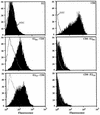
Cell surface expression of chimeric proteins analyzed by flow cytometry. HeLa cells were coinfected with vTF7-3 and the appropriate vaccinia virus recombinant at a multiplicity of infection of 5 PFU/cell. At 8 h postinfection, cells were immunostained with anti-E2 (E2, E2661-CD4, and E2717-CD4) MAb H53 followed by FITC-conjugated rabbit anti-mouse immunoglobulin or with FITC-conjugated anti-CD4 MAb 13B8.2 (CD4, CD4-E2662, and CD4-E2718). Stained cells were fixed in PBS–1% paraformaldehyde before flow cytometric analysis. The level of cell surface expression is indicated by the shift of the solid histogram (MAb H53 plus FITC conjugate or MAb 13B8.2) to the right from the open control histogram (FITC conjugate alone for E2, E2661-CD4, and E2717-CD4; MAb 13B8.2 on uninfected cells for CD4, CD4-E2662, and CD4-E2718).
FIG. 8
Expression of E2, E2661-CD4, and E2717-CD4 analyzed in pulse-chase experiments. HepG2 cells were coinfected with vTF7-3 and the appropriate vaccinia virus recombinant at a multiplicity of infection of 5 PFU/cell. Infected cells were pulse-labeled for 10 min and chased for the indicated times (in hours). Cell lysates were immunoprecipitated with MAb H53 (anti-E2). Samples were separated by SDS-PAGE (10% polyacrylamide). The sizes (in kilodaltons) of protein molecular mass markers are indicated on the right.
FIG. 9
Sensitivities of E2661-CD4 and E2717-CD4 to endo H treatment. HepG2 cells were coinfected with vTF7-3 and the appropriate vaccinia virus recombinant at a multiplicity of infection of 5 PFU/cell. Infected cells were pulse-labeled for 10 min and chased for the indicated times (in hours). Cell lysates were immunoprecipitated with MAb H53 and then treated or not with endo H. Samples were separated by SDS-PAGE (10% polyacrylamide). Deglycosylated proteins are indicated by an asterisk, and endo H-resistant forms are indicated by an “R.” The sizes (in kilodaltons) of protein molecular mass markers are indicated on the right.
FIG. 10
Sensitivities of E2661-GPI and E2717-GPI to PI-PLC treatment. HeLa cells coinfected with vTF7-3 and the appropriate vaccinia virus recombinant were labeled from 4 to 20 h postinfection. Cells were then washed and incubated in PBS in the presence (+) or absence (−) of PI-PLC for 1 h at 37°C. Supernatants and cell pellets were harvested separately and prepared for immunoprecipitation with MAb H53. Samples were separated by SDS-PAGE (10% polyacrylamide). The sizes (in kilodaltons) of protein molecular mass markers are indicated on the right.
FIG. 11
Effect of E2 TMD replacement on the formation of E1-E2 complexes. HepG2 cells were coinfected with vTF7-3, vHCV1-383 (expressing E1), and the appropriate vaccinia virus recombinant at a multiplicity of infection of 5 PFU/cell. Infected cells were labeled from 4 to 20 h postinfection. Cell lysates were immunoprecipitated with MAb H53. Samples were separated by SDS-PAGE (10% polyacrylamide). The sizes (in kilodaltons) of protein molecular mass markers are indicated on the right.
Similar articles
- The transmembrane domain of hepatitis C virus glycoprotein E1 is a signal for static retention in the endoplasmic reticulum.
Cocquerel L, Duvet S, Meunier JC, Pillez A, Cacan R, Wychowski C, Dubuisson J. Cocquerel L, et al. J Virol. 1999 Apr;73(4):2641-9. doi: 10.1128/JVI.73.4.2641-2649.1999. J Virol. 1999. PMID: 10074109 Free PMC article. - Characterization of an endoplasmic reticulum retention signal in the rubella virus E1 glycoprotein.
Hobman TC, Lemon HF, Jewell K. Hobman TC, et al. J Virol. 1997 Oct;71(10):7670-80. doi: 10.1128/JVI.71.10.7670-7680.1997. J Virol. 1997. PMID: 9311850 Free PMC article. - The C-terminal region of the hepatitis C virus E1 glycoprotein confers localization within the endoplasmic reticulum.
Flint M, McKeating JA. Flint M, et al. J Gen Virol. 1999 Aug;80 ( Pt 8):1943-1947. doi: 10.1099/0022-1317-80-8-1943. J Gen Virol. 1999. PMID: 10466789 - Topology of hepatitis C virus envelope glycoproteins.
Op De Beeck A, Dubuisson J. Op De Beeck A, et al. Rev Med Virol. 2003 Jul-Aug;13(4):233-41. doi: 10.1002/rmv.391. Rev Med Virol. 2003. PMID: 12820185 Review. - [Role of N-linked glycans in the functions of hepatitis C virus envelope glycoproteins].
Goffard A, Lazrek M, Bocket L, Dewilde A, Hober D. Goffard A, et al. Ann Biol Clin (Paris). 2007 May-Jun;65(3):237-46. Ann Biol Clin (Paris). 2007. PMID: 17502294 Review. French.
Cited by
- Influence of glycosylation on the immunogenicity and antigenicity of viral immunogens.
Newby ML, Allen JD, Crispin M. Newby ML, et al. Biotechnol Adv. 2024 Jan-Feb;70:108283. doi: 10.1016/j.biotechadv.2023.108283. Epub 2023 Nov 14. Biotechnol Adv. 2024. PMID: 37972669 Free PMC article. Review. - Recognition of native hepatitis C virus E1E2 heterodimers by a human monoclonal antibody.
Cocquerel L, Quinn ER, Flint M, Hadlock KG, Foung SK, Levy S. Cocquerel L, et al. J Virol. 2003 Jan;77(2):1604-9. doi: 10.1128/jvi.77.2.1604-1609.2003. J Virol. 2003. PMID: 12502876 Free PMC article. - The transmembrane domain of the molecular chaperone Cosmc directs its localization to the endoplasmic reticulum.
Sun Q, Ju T, Cummings RD. Sun Q, et al. J Biol Chem. 2011 Apr 1;286(13):11529-42. doi: 10.1074/jbc.M110.173591. Epub 2011 Jan 24. J Biol Chem. 2011. PMID: 21262965 Free PMC article. - Incorporation of hepatitis C virus E1 and E2 glycoproteins: the keystones on a peculiar virion.
Vieyres G, Dubuisson J, Pietschmann T. Vieyres G, et al. Viruses. 2014 Mar 11;6(3):1149-87. doi: 10.3390/v6031149. Viruses. 2014. PMID: 24618856 Free PMC article. Review. - The hepatitis C virus E1 glycoprotein undergoes productive folding but accelerated degradation when expressed as an individual subunit in CHO cells.
Botti V, Bianchi A, Foung SK, Merola M. Botti V, et al. PLoS One. 2011;6(8):e23838. doi: 10.1371/journal.pone.0023838. Epub 2011 Aug 17. PLoS One. 2011. PMID: 21858229 Free PMC article.
References
- Ahn K, Szczesna-Skorupa E, Kemper B. The amino-terminal 29 amino acids of cytochrome P450 2C1 are sufficient for retention in the endoplasmic reticulum. J Biol Chem. 1993;268:18726–18733. - PubMed
- Armstrong J, Patel S. The Golgi sorting domain of coronavirus E1 protein. J Cell Sci. 1991;98:567–575. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials