Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms - PubMed (original) (raw)
Comparative Study
. 1998 Mar 17;95(6):3140-5.
doi: 10.1073/pnas.95.6.3140.
J A Bygraves, E Feil, G Morelli, J E Russell, R Urwin, Q Zhang, J Zhou, K Zurth, D A Caugant, I M Feavers, M Achtman, B G Spratt
Affiliations
- PMID: 9501229
- PMCID: PMC19708
- DOI: 10.1073/pnas.95.6.3140
Comparative Study
Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms
M C Maiden et al. Proc Natl Acad Sci U S A. 1998.
Abstract
Traditional and molecular typing schemes for the characterization of pathogenic microorganisms are poorly portable because they index variation that is difficult to compare among laboratories. To overcome these problems, we propose multilocus sequence typing (MLST), which exploits the unambiguous nature and electronic portability of nucleotide sequence data for the characterization of microorganisms. To evaluate MLST, we determined the sequences of approximately 470-bp fragments from 11 housekeeping genes in a reference set of 107 isolates of Neisseria meningitidis from invasive disease and healthy carriers. For each locus, alleles were assigned arbitrary numbers and dendrograms were constructed from the pairwise differences in multilocus allelic profiles by cluster analysis. The strain associations obtained were consistent with clonal groupings previously determined by multilocus enzyme electrophoresis. A subset of six gene fragments was chosen that retained the resolution and congruence achieved by using all 11 loci. Most isolates from hyper-virulent lineages of serogroups A, B, and C meningococci were identical for all loci or differed from the majority type at only a single locus. MLST using six loci therefore reliably identified the major meningococcal lineages associated with invasive disease. MLST can be applied to almost all bacterial species and other haploid organisms, including those that are difficult to cultivate. The overwhelming advantage of MLST over other molecular typing methods is that sequence data are truly portable between laboratories, permitting one expanding global database per species to be placed on a World-Wide Web site, thus enabling exchange of molecular typing data for global epidemiology via the Internet.
Figures
Figure 1
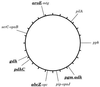
Chromosomal locations of gene fragments. The locations are drawn on the physical map of strain Z2491 (12), a subgroup IV-1 strain. The six loci chosen for MLST are shown in boldfaced, underlined text. aroE and mtg are located next to each other (14) on _Bgl_II fragment B14 (41 kb). pip and opaJ are also next to each other (13) (data not shown) and are located on _Bgl_II fragment B16 (32 kb). serC and opaB are located within a few kilobases of each other (13) as are abcZ and opc (unpublished data). pgm and adk hybridized to the same set of fragments, including B7 and P3, which overlap by ≈50 kb. gdh mapped on _Spe_I fragment S17 (35 kb).
Figure 2
Dendrogram of genetic relationships among 107 strains based on 6 gene fragments. Linkage distance is indicated by a scale at the top, and the MLEE or ST assignments of lineages are indicated by shaded rectangles. The asterisk indicates ST-21 (serogroup A strain B534).
Similar articles
- Characterization of Neisseria meningitidis isolates collected from 1974 to 2003 in Japan by multilocus sequence typing.
Takahashi H, Kuroki T, Watanabe Y, Tanaka H, Inouye H, Yamai S, Watanabe H. Takahashi H, et al. J Med Microbiol. 2004 Jul;53(Pt 7):657-662. doi: 10.1099/jmm.0.45541-0. J Med Microbiol. 2004. PMID: 15184538 - Molecular typing methods for Neisseria meningitidis.
Yakubu DE, Abadi FJR, Pennington TH. Yakubu DE, et al. J Med Microbiol. 1999 Dec;48(12):1055-1064. doi: 10.1099/00222615-48-12-1055. J Med Microbiol. 1999. PMID: 10591158 Review. - Semiautomation of multilocus sequence typing for the characterization of clinical isolates of Neisseria meningitidis.
Clarke SC, Diggle MA, Edwards GF. Clarke SC, et al. J Clin Microbiol. 2001 Sep;39(9):3066-71. doi: 10.1128/JCM.39.9.3066-3071.2001. J Clin Microbiol. 2001. PMID: 11526130 Free PMC article. - Nucleotide sequence-based typing of meningococci directly from clinical samples.
Diggle MA, Bell CM, Clarke SC. Diggle MA, et al. J Med Microbiol. 2003 Jun;52(Pt 6):505-508. doi: 10.1099/jmm.0.05078-0. J Med Microbiol. 2003. PMID: 12748270 - Multilocus sequence typing of bacteria.
Maiden MC. Maiden MC. Annu Rev Microbiol. 2006;60:561-88. doi: 10.1146/annurev.micro.59.030804.121325. Annu Rev Microbiol. 2006. PMID: 16774461 Review.
Cited by
- STing: accurate and ultrafast genomic profiling with exact sequence matches.
Espitia-Navarro HF, Chande AT, Nagar SD, Smith H, Jordan IK, Rishishwar L. Espitia-Navarro HF, et al. Nucleic Acids Res. 2020 Aug 20;48(14):7681-7689. doi: 10.1093/nar/gkaa566. Nucleic Acids Res. 2020. PMID: 32619234 Free PMC article. - Not Only Meningitis but Also Epiglottitis: An Emerging Clinical Presentation of Invasive Meningococcal Disease.
Deghmane AE, Taha S, Taha MK. Deghmane AE, et al. Open Forum Infect Dis. 2023 Dec 8;11(1):ofad615. doi: 10.1093/ofid/ofad615. eCollection 2024 Jan. Open Forum Infect Dis. 2023. PMID: 38192380 Free PMC article. - Resolution of a Protracted Serogroup B Meningococcal Outbreak with Whole-Genome Sequencing Shows Interspecies Genetic Transfer.
Mulhall RM, Brehony C, O'Connor L, Meyler K, Jolley KA, Bray J, Bennett D, Maiden MC, Cunney R. Mulhall RM, et al. J Clin Microbiol. 2016 Dec;54(12):2891-2899. doi: 10.1128/JCM.00881-16. Epub 2016 Sep 14. J Clin Microbiol. 2016. PMID: 27629899 Free PMC article. - Whole-Genome Multilocus Sequence Typing of Extended-Spectrum-Beta-Lactamase-Producing Enterobacteriaceae.
Kluytmans-van den Bergh MF, Rossen JW, Bruijning-Verhagen PC, Bonten MJ, Friedrich AW, Vandenbroucke-Grauls CM, Willems RJ, Kluytmans JA. Kluytmans-van den Bergh MF, et al. J Clin Microbiol. 2016 Dec;54(12):2919-2927. doi: 10.1128/JCM.01648-16. Epub 2016 Sep 14. J Clin Microbiol. 2016. PMID: 27629900 Free PMC article. - Predictive diagnostics for Escherichia coli infections based on the clonal association of antimicrobial resistance and clinical outcome.
Tchesnokova V, Billig M, Chattopadhyay S, Linardopoulou E, Aprikian P, Roberts PL, Skrivankova V, Johnston B, Gileva A, Igusheva I, Toland A, Riddell K, Rogers P, Qin X, Butler-Wu S, Cookson BT, Fang FC, Kahl B, Price LB, Weissman SJ, Limaye A, Scholes D, Johnson JR, Sokurenko EV. Tchesnokova V, et al. J Clin Microbiol. 2013 Sep;51(9):2991-9. doi: 10.1128/JCM.00984-13. Epub 2013 Jul 10. J Clin Microbiol. 2013. PMID: 23843485 Free PMC article.
References
- Achtman M. In: Molecular Medical Microbiology. Sussman M, editor. London: Academic; 1998. , in press.
- Selander R K, Musser J M, Caugant D A, Gilmour M N, Whittam T S. Microb Pathog. 1987;3:1–7. - PubMed
- Caugant D A, Bøvre K, Gaustad P, Bryn K, Holten E, Høiby E A, Frøholm L O. J Gen Microbiol. 1986;132:641–652. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical