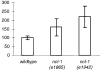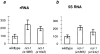ncl-1 is required for the regulation of cell size and ribosomal RNA synthesis in Caenorhabditis elegans - PubMed (original) (raw)
ncl-1 is required for the regulation of cell size and ribosomal RNA synthesis in Caenorhabditis elegans
D J Frank et al. J Cell Biol. 1998.
Abstract
Regulation of ribosome synthesis is an essential aspect of growth control. Thus far, little is known about the factors that control and coordinate these processes. We show here that the Caenorhabditis elegans gene ncl-1 encodes a zinc finger protein and may be a repressor of RNA polymerase I and III transcription and an inhibitor of cell growth. Loss of function mutations in ncl-1, previously shown to result in enlarged nucleoli, result in increased rates of rRNA and 5S RNA transcription and enlarged cells. Furthermore, ncl-1 adult worms are larger, have more protein, and have twice as much rRNA as wild-type worms. Localization studies show that the level of NCL-1 protein is independently regulated in different cells of the embryo. In wild-type embryos, cells with the largest nucleoli have the lowest level of NCL-1 protein. Based on these results we propose that ncl-1 is a repressor of ribosome synthesis and cell growth.
Figures
Figure 3
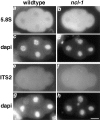
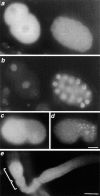
Increased nuclear precursor rRNA in ncl-1 mutants. Wild-type (a, c, e, and g) and ncl-1 (b, d, f, and h) embryos were hybridized to 5.8S (a and b) or ITS2 (e and f) digoxigenin-labeled antisense oligo probes. Probes were visualized using an antidigoxigenin antibody coupled to FITC. DNA was visualized by staining with 4′,6-diamidino-2-phenylindole (DAPI) (c, d, g, and h correspond to a, b, e, and f respectively). In ncl-1, nucleoli are detectable with both the 5.8S probe (b) which detects both precursor and mature rRNA, and the ITS2 probe (f) which detects only unprocessed precursor rRNA. Bar, 10 μm.
Figure 1
Nucleoli are visible in ncl-1 but not wild-type early embryos. Live wild-type and ncl-1 four-cell embryos were visualized using differential interference contrast microscopy. One or two nucleoli can be seen in all nuclei of ncl-1 embryos; in this image nucleoli in the most anterior blastomere are out of the plane of focus. The nucleolus in the most posterior blastomere is indicated with an arrow. In this and all other figures, anterior is to the left and dorsal is up. Bar, 10 μm.
Figure 2
Increased steady-state level of rRNA in ncl-1 mutants. RNA was isolated from three samples each of wild-type, ncl-1 (e1865), and ncl-1 (e1942) worms grown at 20°C for 48 h after hatching. RNA was run on a 1% agarose formaldehyde gel and then transferred to nitrocellulose. The same blot was hybridized with both histone and 28S rRNA probes. The graph represents relative levels of 28S rRNA standardized to histone message. Quantitation was performed with a phosphorimager. This experiment was performed twice with similar outcomes; the data from one experiment is presented here. Error bars represent standard deviations.
Figure 4
Increased rate of rRNA and 5S RNA transcription in ncl-1 mutants. Equivalent numbers of nuclei from wild-type, ncl-1 (e1865), and ncl-1 (e1942) early embryos were labeled in nuclear run-on assays (refer to Materials and Methods). For each reaction, radiolabeled RNA was extracted and then hybridized to a slot blot filter containing: (a) one repeat of rDNA; (b) one copy of 5S RNA; or (c) one copy each of histones H2A, H2B, H3, and H4. In each case, graphs represent a minimum of two independent nuclei preparations and three reactions per preparation. The y axis represents percentages with wild-type set to 100. Quantitation was performed with a phosphorimager. Error bars represent standard deviations. The probability (P) that the mutants are different than wild-type is as follows: ncl-1 (e1865) rRNA, P < 0.005; ncl-1 (e1942) rRNA, P < 0.02; ncl-1 (e1865) 5S RNA, P < 0.001; ncl-1 (e1942) 5S RNA, P < 0.2; ncl-1 (e1865) histone, P < 0.5; ncl-1 (e1942), P < 0.5.
Figure 4
Increased rate of rRNA and 5S RNA transcription in ncl-1 mutants. Equivalent numbers of nuclei from wild-type, ncl-1 (e1865), and ncl-1 (e1942) early embryos were labeled in nuclear run-on assays (refer to Materials and Methods). For each reaction, radiolabeled RNA was extracted and then hybridized to a slot blot filter containing: (a) one repeat of rDNA; (b) one copy of 5S RNA; or (c) one copy each of histones H2A, H2B, H3, and H4. In each case, graphs represent a minimum of two independent nuclei preparations and three reactions per preparation. The y axis represents percentages with wild-type set to 100. Quantitation was performed with a phosphorimager. Error bars represent standard deviations. The probability (P) that the mutants are different than wild-type is as follows: ncl-1 (e1865) rRNA, P < 0.005; ncl-1 (e1942) rRNA, P < 0.02; ncl-1 (e1865) 5S RNA, P < 0.001; ncl-1 (e1942) 5S RNA, P < 0.2; ncl-1 (e1865) histone, P < 0.5; ncl-1 (e1942), P < 0.5.
Figure 5
Rescue and antisense phenocopy of ncl-1. Each image shows a region of a worm anterior of the posterior bulb of the pharynx. Nuclei of neurons in this region are indicated with arrows. In wild type, these nuclei have very small or no detectable nucleoli. In ncl-1 worms, neuronal nuclei have large nucleoli. Injection of a 7.5-kb fragment of genomic DNA containing the ncl-1 gene into the syncytial gonad of ncl-1 worms results in partially rescued F1 progeny (ncl-1 rescue). These worms have neurons that do not have large nucleoli. Injection of antisense RNA from the ncl-1 gene into the syncytial gonad of wild-type worms results in phenocopy in F1 progeny (WT antisense). Neurons in these wild-type worms have enlarged nucleoli. Bar, 10 μm.
Figure 6
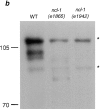
Predicted amino acid sequence and Western blot analysis of NCL-1. (a) A ncl-1 cDNA cloned by hybridization to the rescuing genomic fragment confirmed the exon structure of the gene ZK112.2 predicted by the C. elegans genome consortium. The 851-amino acid open reading frame contains two B box zinc finger motifs (amino acids 132–164 and 223–256) that are shaded with conserved cysteines and histidines in bold. Predicted coiled-coil motif (amino acids 257–363, determined using the Matcher program) is underlined. The predicted molecular weight of NCL-1 is 92 kD. (b) Extracts of wild-type, ncl-1 (e1865), and ncl-1 (e1942) embryos were run on a 6% SDS-PAGE gel, transferred to nitrocellulose, and then probed with mAb D3C2. Background bands detected without primary antibody are denoted by asterisks. mAb D3C2 recognizes two bands of ∼97-kD in wild-type but not ncl-1 extracts. No immunoreactive species is detected in extracts from ncl-1(e1942), whereas a faster migrating species of ∼80-kD is detected in extracts from ncl-1 (e1865) embryos.
Figure 6
Predicted amino acid sequence and Western blot analysis of NCL-1. (a) A ncl-1 cDNA cloned by hybridization to the rescuing genomic fragment confirmed the exon structure of the gene ZK112.2 predicted by the C. elegans genome consortium. The 851-amino acid open reading frame contains two B box zinc finger motifs (amino acids 132–164 and 223–256) that are shaded with conserved cysteines and histidines in bold. Predicted coiled-coil motif (amino acids 257–363, determined using the Matcher program) is underlined. The predicted molecular weight of NCL-1 is 92 kD. (b) Extracts of wild-type, ncl-1 (e1865), and ncl-1 (e1942) embryos were run on a 6% SDS-PAGE gel, transferred to nitrocellulose, and then probed with mAb D3C2. Background bands detected without primary antibody are denoted by asterisks. mAb D3C2 recognizes two bands of ∼97-kD in wild-type but not ncl-1 extracts. No immunoreactive species is detected in extracts from ncl-1(e1942), whereas a faster migrating species of ∼80-kD is detected in extracts from ncl-1 (e1865) embryos.
Figure 7
Localization of NCL-1 protein. (a and b) Immunofluorescence micrographs of wild-type embryos stained for NCL-1 protein with mAb D3C2 (a) and with DAPI to visualize nuclei (b). Intense staining is seen in the two-cell embryo (left), but is diminished by the 28-cell stage (right). (c and d) Immunofluorescence micrographs of two different wild-type 300-cell embryos stained with either mAb D3C2 to visualize NCL-1 protein (c) or with mAb K121 (d) to visualize nucleoli. At this stage, NCL-1 protein appears to be present in all cells except for those of the gut which have prominent nucleoli as detected by mAb K121. (e) Immunofluorescence micrograph of a wild-type gonad stained with mAb D3C2. Staining is most intense in the most mature oocyte (left). The large and small brackets indicate the most mature and second most mature oocytes, respectively. Bars, 10 μm.
Similar articles
- A Genetic Cascade of let-7-ncl-1-fib-1 Modulates Nucleolar Size and rRNA Pool in Caenorhabditis elegans.
Yi YH, Ma TH, Lee LW, Chiou PT, Chen PH, Lee CM, Chu YD, Yu H, Hsiung KC, Tsai YT, Lee CC, Chang YS, Chan SP, Tan BC, Lo SJ. Yi YH, et al. PLoS Genet. 2015 Oct 22;11(10):e1005580. doi: 10.1371/journal.pgen.1005580. eCollection 2015 Oct. PLoS Genet. 2015. PMID: 26492166 Free PMC article. - The Drosophila melanogaster gene brain tumor negatively regulates cell growth and ribosomal RNA synthesis.
Frank DJ, Edgar BA, Roth MB. Frank DJ, et al. Development. 2002 Jan;129(2):399-407. doi: 10.1242/dev.129.2.399. Development. 2002. PMID: 11807032 - The ncl-1 gene and genetic mosaics of Caenorhabditis elegans.
Hedgecock EM, Herman RK. Hedgecock EM, et al. Genetics. 1995 Nov;141(3):989-1006. doi: 10.1093/genetics/141.3.989. Genetics. 1995. PMID: 8582642 Free PMC article. - Control points in eucaryotic ribosome biogenesis.
Larson DE, Zahradka P, Sells BH. Larson DE, et al. Biochem Cell Biol. 1991 Jan;69(1):5-22. doi: 10.1139/o91-002. Biochem Cell Biol. 1991. PMID: 2043343 Review. - Eukaryotic 5S rRNA biogenesis.
Ciganda M, Williams N. Ciganda M, et al. Wiley Interdiscip Rev RNA. 2011 Jul-Aug;2(4):523-33. doi: 10.1002/wrna.74. Epub 2011 Feb 25. Wiley Interdiscip Rev RNA. 2011. PMID: 21957041 Free PMC article. Review.
Cited by
- Fourier transform infrared microspectroscopy for the analysis of the biochemical composition of C. elegans worms.
Sheng M, Gorzsás A, Tuck S. Sheng M, et al. Worm. 2016 Feb 18;5(1):e1132978. doi: 10.1080/21624054.2015.1132978. eCollection 2016 Jan-Mar. Worm. 2016. PMID: 27073735 Free PMC article. - Differential contributions of Caenorhabditis elegans histone deacetylases to huntingtin polyglutamine toxicity.
Bates EA, Victor M, Jones AK, Shi Y, Hart AC. Bates EA, et al. J Neurosci. 2006 Mar 8;26(10):2830-8. doi: 10.1523/JNEUROSCI.3344-05.2006. J Neurosci. 2006. PMID: 16525063 Free PMC article. - Control of asymmetric cell division in early C. elegans embryogenesis: teaming-up translational repression and protein degradation.
Hwang SY, Rose LS. Hwang SY, et al. BMB Rep. 2010 Feb;43(2):69-78. doi: 10.5483/bmbrep.2010.43.2.069. BMB Rep. 2010. PMID: 20193124 Free PMC article. Review. - The tripartite motif family identifies cell compartments.
Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, Riganelli D, Zanaria E, Messali S, Cainarca S, Guffanti A, Minucci S, Pelicci PG, Ballabio A. Reymond A, et al. EMBO J. 2001 May 1;20(9):2140-51. doi: 10.1093/emboj/20.9.2140. EMBO J. 2001. PMID: 11331580 Free PMC article. - A Genetic Cascade of let-7-ncl-1-fib-1 Modulates Nucleolar Size and rRNA Pool in Caenorhabditis elegans.
Yi YH, Ma TH, Lee LW, Chiou PT, Chen PH, Lee CM, Chu YD, Yu H, Hsiung KC, Tsai YT, Lee CC, Chang YS, Chan SP, Tan BC, Lo SJ. Yi YH, et al. PLoS Genet. 2015 Oct 22;11(10):e1005580. doi: 10.1371/journal.pgen.1005580. eCollection 2015 Oct. PLoS Genet. 2015. PMID: 26492166 Free PMC article.
References
- Albertson DG. Formation of the first cleavage spindle in nematode embryos. Dev Biol. 1984;101:61–72. - PubMed
- Allo SN, McDermott PJ, Carl LL, Morgan HE. Phorbol ester stimulation of protein kinase C activity and ribosomal DNA transcription. J Biol Chem. 1991;266:22003–22009. - PubMed
- Altman, P.L., and D.D. Katz 1976. Cell Biology. FASEB (Fed. Am. Soc. Exp. Biol.), Bethesda, MD. 454 pp.
- Altmann GG, Leblond CP. Changes in the size and structure of the nucleolus of columnar cells during their migration from crypt base to villus top in rat jejunum. J Cell Sci. 1982;56:83–99. - PubMed
- Ausubel, F.M., R. Brent, R.R. Kingston, D.D. Moore, J.G. Seideman, J.A. Smith, and K. Struhl. 1987. Analysis of RNA by Northern Hybridization. In Current Protocols in Molecular Biology. Vol. 1. Wiley/Greene, New York. 4.9.1–4.9.4.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous