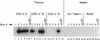Helios, a T cell-restricted Ikaros family member that quantitatively associates with Ikaros at centromeric heterochromatin - PubMed (original) (raw)
Helios, a T cell-restricted Ikaros family member that quantitatively associates with Ikaros at centromeric heterochromatin
K Hahm et al. Genes Dev. 1998.
Abstract
The Ikaros gene encodes multiple protein isoforms that contribute critical functions during the development of lymphocytes and other hematopoietic cell types. The intracellular functions of Ikaros are not known, although recent studies have shown that Ikaros proteins colocalize with inactive genes and centromeric heterochromatin. In this study, Ikaros proteins were found to be components of highly stable complexes. The complexes from an immature T cell line were purified, revealing associated proteins of 70 and 30 kD. The p70 gene, named Helios, encodes two protein isoforms with zinc finger domains exhibiting considerable homology to those within Ikaros proteins. Helios and Ikaros recognize similar DNA sequences and, when overexpressed, Helios associates indiscriminately with the various Ikaros isoforms. Although Ikaros is present in most hematopoietic cells, Helios was found primarily in T cells. The relevance of the Ikaros-Helios interaction in T cells is supported by the quantitative association of Helios with a fraction of the Ikaros. Interestingly, the Ikaros-Helios complexes localize to the centromeric regions of T cell nuclei, similar to the Ikaros localization previously observed in B cells. Unlike the B cell results, however, only a fraction of the Ikaros, presumably the fraction associated with Helios, exhibited centromeric localization in T cells. These results establish immunoaffinity chromatography as a useful method for identifying Ikaros partners and suggest that Helios is a limiting regulatory subunit for Ikaros within centromeric heterochromatin.
Figures
Figure 1
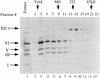
Coelution of Ikaros isoforms in a broad peak from gel filtration columns. RLm11 nuclear extracts (1 mg) were analyzed by Superdex 200 (Pharmacia) gel filtration chromatography. Five micrograms of nuclear extract (lane 1) and 45 μl of every other column fraction (lanes 2–13) were separated by SDS-PAGE and analyzed by immunoblotting, with antiserum directed against Ikaros and Elf-1. The fractions in which standard molecular mass markers elute are indicated by arrows at the top, and the bands corresponding to Elf-1 and Ikaros isoforms I, III, V, and VI are (left).
Figure 2
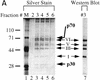
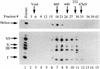
Immunoaffinity purification of Ikaros complexes from RLm11 nuclear extracts. (A) RLm11 nuclear extracts were applied to a protein A–Sepharose column containing covalently linked antibodies directed against the carboxy-terminal half of Ikaros (see Materials and Methods). After washing the resin with buffers containing 0.45 and 1
m
KCl, bound proteins were eluted with 100 m
m
trimethyl ethanolamine (pH 11.0). The trimethyl ethanolamine fractions (numbers 2 through 6, lanes 2–6) were analyzed by SDS-PAGE followed by silver staining. Molecular mass markers are shown in lane 1 and are indicated to the left. Fraction 3 was also analyzed by immunoblotting with anti-Ikaros serum (lane 7). Ikaros isoforms I, III, V, and VI are indicated between lanes 6 and 7. Two proteins, p30 and p70, were detected by silver staining that did not interact with the Ikaros antibodies. (B) RLm11 extracts were applied to protein A–Sepharose columns containing covalently linked antibodies directed against Elf-1 (lane 2), the carboxyl-terminus of Ikaros (lane 3), or the amino-terminus of Ikaros (lane 4). The columns were washed and proteins eluted as described above. Portions of the trimethyl ethanolamine eluates were analyzed by SDS-PAGE followed by silver-staining. Also analyzed were proteins purified by sequence-specific DNA-affinity chromatography with a resin containing covalently linked multimers of the TdT D element (lane 5, see Materials and Methods and Hahm et al. 1994). Molecular mass markers are shown in lane 1 and are indicated at left in kD. Ikaros isoforms I, III, V, and VI, p70, and p30, are indicated at right.
Figure 2
Immunoaffinity purification of Ikaros complexes from RLm11 nuclear extracts. (A) RLm11 nuclear extracts were applied to a protein A–Sepharose column containing covalently linked antibodies directed against the carboxy-terminal half of Ikaros (see Materials and Methods). After washing the resin with buffers containing 0.45 and 1
m
KCl, bound proteins were eluted with 100 m
m
trimethyl ethanolamine (pH 11.0). The trimethyl ethanolamine fractions (numbers 2 through 6, lanes 2–6) were analyzed by SDS-PAGE followed by silver staining. Molecular mass markers are shown in lane 1 and are indicated to the left. Fraction 3 was also analyzed by immunoblotting with anti-Ikaros serum (lane 7). Ikaros isoforms I, III, V, and VI are indicated between lanes 6 and 7. Two proteins, p30 and p70, were detected by silver staining that did not interact with the Ikaros antibodies. (B) RLm11 extracts were applied to protein A–Sepharose columns containing covalently linked antibodies directed against Elf-1 (lane 2), the carboxyl-terminus of Ikaros (lane 3), or the amino-terminus of Ikaros (lane 4). The columns were washed and proteins eluted as described above. Portions of the trimethyl ethanolamine eluates were analyzed by SDS-PAGE followed by silver-staining. Also analyzed were proteins purified by sequence-specific DNA-affinity chromatography with a resin containing covalently linked multimers of the TdT D element (lane 5, see Materials and Methods and Hahm et al. 1994). Molecular mass markers are shown in lane 1 and are indicated at left in kD. Ikaros isoforms I, III, V, and VI, p70, and p30, are indicated at right.
Figure 3
Helios A and Helios B contain zinc fingers with homology to those in Ikaros and Aiolos. An amino acid sequence alignment compiled by use of the PILEUP program is shown. Compared are the largest isoforms of the Ikaros family members, Helios B, Ikaros isoform VI (Ik-1), and Aiolos. Lowercase lettering in the Helios B sequence designates the sole difference between Helios B and Helios A. Amino acid sequences obtained by microsequencing of purified Helios protein are indicated by a line above the Helios sequence. Shaded boxes in blue emphasize the highly conserved zinc finger motifs. Yellow bars indicate the conserved cysteines and histidines in the zinc fingers. The red box indicates the unusual cysteine in the apparent C2HC finger. Overall sequence similarities are as follows: Helios B–Ikaros, 55%; Ikaros–Aiolos, 53%; Helios B–Aiolos, 50%. The four amino-terminal zinc fingers of Helios share 94% identity with the Ikaros fingers and the Aiolos fingers share 86% identity with the Ikaros fingers. The two carboxy-terminal zinc fingers of Helios share 85% identity with the corresponding Ikaros fingers, with 80% identity between the Aiolos and Ikaros fingers. The Helios cDNA and amino acid sequences have been deposited in the GenBank/Swiss Prot databases (accession nos. AF044257 and P81183, respectively).
Figure 4
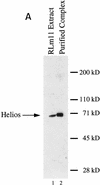
Indiscriminate interactions between Helios A and Ikaros isoforms in 293T cells. (A) The Helios protein within an RLm11 extract (lane 1) and the immunoaffinity purified Ikaros complex (lane 2) can be detected by immunoblot analysis with antisera directed against recombinant Helios (amino acids 1–109). Molecular mass markers are indicated at right, and the location of the Helios band is indicated at left. (B) 293T cells were transfected with 10 or 15 μg of expression plasmids for various Ikaros isoforms (lanes 1–18; specific isoforms indicated above each lane), in the absence (lanes 1,2) or presence (lanes 3–18) of an expression plasmid for FLAG-tagged Helios A (5 μg). Cytoplasmic (odd-numbered lanes) and nuclear (even numbered lanes) extracts from the transfected cells were analyzed by immunoblotting with antibodies directed against both Helios and Ikaros. The bands corresponding to FLAG-tagged Helios A and Ikaros isoforms I, V, and VI are indicated to the left. (C) Interactions between Helios A and Ikaros isoforms were assessed by immunoprecipitation from the nuclear extracts shown in part B with a monoclonal antibody directed against the FLAG epitope. Proteins within the immunoprecipitated pellet were analyzed by immunoblotting with antisera directed against Helios (top) or Ikaros (bottom). The extacts used for immunoprecipitation contained (lanes 3–10) or lacked (lanes 1,2) the FLAG–Helios A protein and zero, one, two, or three Ikaros isoforms (isoforms indicated above each lane). The locations of the bands corresponding to FLAG–Helios A and Ikaros isoforms I, V, and VI are indicated to the right.
Figure 4
Indiscriminate interactions between Helios A and Ikaros isoforms in 293T cells. (A) The Helios protein within an RLm11 extract (lane 1) and the immunoaffinity purified Ikaros complex (lane 2) can be detected by immunoblot analysis with antisera directed against recombinant Helios (amino acids 1–109). Molecular mass markers are indicated at right, and the location of the Helios band is indicated at left. (B) 293T cells were transfected with 10 or 15 μg of expression plasmids for various Ikaros isoforms (lanes 1–18; specific isoforms indicated above each lane), in the absence (lanes 1,2) or presence (lanes 3–18) of an expression plasmid for FLAG-tagged Helios A (5 μg). Cytoplasmic (odd-numbered lanes) and nuclear (even numbered lanes) extracts from the transfected cells were analyzed by immunoblotting with antibodies directed against both Helios and Ikaros. The bands corresponding to FLAG-tagged Helios A and Ikaros isoforms I, V, and VI are indicated to the left. (C) Interactions between Helios A and Ikaros isoforms were assessed by immunoprecipitation from the nuclear extracts shown in part B with a monoclonal antibody directed against the FLAG epitope. Proteins within the immunoprecipitated pellet were analyzed by immunoblotting with antisera directed against Helios (top) or Ikaros (bottom). The extacts used for immunoprecipitation contained (lanes 3–10) or lacked (lanes 1,2) the FLAG–Helios A protein and zero, one, two, or three Ikaros isoforms (isoforms indicated above each lane). The locations of the bands corresponding to FLAG–Helios A and Ikaros isoforms I, V, and VI are indicated to the right.
Figure 4
Indiscriminate interactions between Helios A and Ikaros isoforms in 293T cells. (A) The Helios protein within an RLm11 extract (lane 1) and the immunoaffinity purified Ikaros complex (lane 2) can be detected by immunoblot analysis with antisera directed against recombinant Helios (amino acids 1–109). Molecular mass markers are indicated at right, and the location of the Helios band is indicated at left. (B) 293T cells were transfected with 10 or 15 μg of expression plasmids for various Ikaros isoforms (lanes 1–18; specific isoforms indicated above each lane), in the absence (lanes 1,2) or presence (lanes 3–18) of an expression plasmid for FLAG-tagged Helios A (5 μg). Cytoplasmic (odd-numbered lanes) and nuclear (even numbered lanes) extracts from the transfected cells were analyzed by immunoblotting with antibodies directed against both Helios and Ikaros. The bands corresponding to FLAG-tagged Helios A and Ikaros isoforms I, V, and VI are indicated to the left. (C) Interactions between Helios A and Ikaros isoforms were assessed by immunoprecipitation from the nuclear extracts shown in part B with a monoclonal antibody directed against the FLAG epitope. Proteins within the immunoprecipitated pellet were analyzed by immunoblotting with antisera directed against Helios (top) or Ikaros (bottom). The extacts used for immunoprecipitation contained (lanes 3–10) or lacked (lanes 1,2) the FLAG–Helios A protein and zero, one, two, or three Ikaros isoforms (isoforms indicated above each lane). The locations of the bands corresponding to FLAG–Helios A and Ikaros isoforms I, V, and VI are indicated to the right.
Figure 5
Helios A and Ikaros isoform V bind to similar DNA sequences. (A) High-affinity Helios A-binding sites were selected by a GST pull-down assay (Zweidler-McKay et al. 1996). DNA fragments selected by the GST–Helios A fusion protein were sequenced and tabulated. The selected sequences can be divided into two groups, both containing a core sequence of GGA. Group 1 was represented by 19 sequences and Group 2, by 30 sequences. The number of fragments containing each nucleotide at a given position following alignment are indicated for each group. Only 14 sequences are shown for the first four positions of the Group 2 sequence because the remaining 16 were derived from fragments in which these four positions were contained within the invariant primer-binding sites flanking the variable sequences. (B) Gel mobility shift assays were performed with recombinant GST–Ikaros isoform V (lanes 1–6) and recombinant Helios A (lanes 7–12) with probes containing the five sequences shown at the bottom (lanes 2–6, 8–12) and a negative control probe (lanes 1,7).The specific probes were derived from the pSP72 (Promega) multiple cloning site region (lanes 1,7), the TdT D (lanes 2,8) and D′ (lanes 3,9) elements, the Hs BS1 (i.e., Helios binding site 1; lanes 4,10) and Hs BS2 (lanes 5,11) sequences selected in this study, and the IkBS2 (lanes 6,12) sequence selected previously (Molnar and Georgopoulos 1994).
Figure 5
Helios A and Ikaros isoform V bind to similar DNA sequences. (A) High-affinity Helios A-binding sites were selected by a GST pull-down assay (Zweidler-McKay et al. 1996). DNA fragments selected by the GST–Helios A fusion protein were sequenced and tabulated. The selected sequences can be divided into two groups, both containing a core sequence of GGA. Group 1 was represented by 19 sequences and Group 2, by 30 sequences. The number of fragments containing each nucleotide at a given position following alignment are indicated for each group. Only 14 sequences are shown for the first four positions of the Group 2 sequence because the remaining 16 were derived from fragments in which these four positions were contained within the invariant primer-binding sites flanking the variable sequences. (B) Gel mobility shift assays were performed with recombinant GST–Ikaros isoform V (lanes 1–6) and recombinant Helios A (lanes 7–12) with probes containing the five sequences shown at the bottom (lanes 2–6, 8–12) and a negative control probe (lanes 1,7).The specific probes were derived from the pSP72 (Promega) multiple cloning site region (lanes 1,7), the TdT D (lanes 2,8) and D′ (lanes 3,9) elements, the Hs BS1 (i.e., Helios binding site 1; lanes 4,10) and Hs BS2 (lanes 5,11) sequences selected in this study, and the IkBS2 (lanes 6,12) sequence selected previously (Molnar and Georgopoulos 1994).
Figure 6
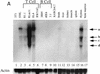
Helios expression is largely restricted in T cells. (A) Northern blot analysis was used to measure Helios mRNA expression in murine hematopoietic cell lines and tissues. Cell lines and tissues analyzed are indicated at the top. Twenty micrograms of total RNAs was analyzed in each lane. Nitrocellulose membranes were hybridized sequentially with a Helios probe (fragment encoding amino acids 221–292, top) and a human β-actin probe (500 bp, bottom). Positions of the 28S and 18S rRNAs are indicated. Actin transcripts and four major transcripts detected by Helios probe are indicated. (B) Helios protein in murine cell lines was analyzed by immunoblotting. Cell lines analyzed are indicated at the top. Twenty micrograms of protein from nuclear extracts estimated by Coomassie Assay (Pierce) were loaded in each lane. (p70) Protein migrating close to 70 kD and detected with anti-Helios serum. (C) The nuclear extracts analyzed in B were probes with antibodies directed against the carboxy-terminal half of Ikaros.
Figure 6
Helios expression is largely restricted in T cells. (A) Northern blot analysis was used to measure Helios mRNA expression in murine hematopoietic cell lines and tissues. Cell lines and tissues analyzed are indicated at the top. Twenty micrograms of total RNAs was analyzed in each lane. Nitrocellulose membranes were hybridized sequentially with a Helios probe (fragment encoding amino acids 221–292, top) and a human β-actin probe (500 bp, bottom). Positions of the 28S and 18S rRNAs are indicated. Actin transcripts and four major transcripts detected by Helios probe are indicated. (B) Helios protein in murine cell lines was analyzed by immunoblotting. Cell lines analyzed are indicated at the top. Twenty micrograms of protein from nuclear extracts estimated by Coomassie Assay (Pierce) were loaded in each lane. (p70) Protein migrating close to 70 kD and detected with anti-Helios serum. (C) The nuclear extracts analyzed in B were probes with antibodies directed against the carboxy-terminal half of Ikaros.
Figure 6
Helios expression is largely restricted in T cells. (A) Northern blot analysis was used to measure Helios mRNA expression in murine hematopoietic cell lines and tissues. Cell lines and tissues analyzed are indicated at the top. Twenty micrograms of total RNAs was analyzed in each lane. Nitrocellulose membranes were hybridized sequentially with a Helios probe (fragment encoding amino acids 221–292, top) and a human β-actin probe (500 bp, bottom). Positions of the 28S and 18S rRNAs are indicated. Actin transcripts and four major transcripts detected by Helios probe are indicated. (B) Helios protein in murine cell lines was analyzed by immunoblotting. Cell lines analyzed are indicated at the top. Twenty micrograms of protein from nuclear extracts estimated by Coomassie Assay (Pierce) were loaded in each lane. (p70) Protein migrating close to 70 kD and detected with anti-Helios serum. (C) The nuclear extracts analyzed in B were probes with antibodies directed against the carboxy-terminal half of Ikaros.
Figure 7
Helios expression in thymocyte and splenic cell subsets. Twenty cells of the indicated cell-surface phenotypes were sorted by FACS for analysis of Helios expression during thymocyte maturation and in non-T-lineage cell subsets. Primers for nested PCR amplified across sequences that encode the four amino-terminal zinc fingers of Helios. The PCR products observed with the various samples exhibit slightly different migrations, which represent the amplification of both the Helios A and Helios B products with predicted sizes of 712 and 790 bp, respectively.
Figure 8
Quantitative association of Helios with a subset of the Ikaros within VL3-3M2 cells. Quantitative immunoprecipitation experiments were performed with purified IgG directed against the amino-terminal domains of either Ikaros (lanes 2–5,11–14) or Helios (lanes 6–9,15–18). Control immunoprecipitations contained no added antibody (lanes 1,10). The proteins present in the immunoprecipitation pellets (lanes 1–9) and supernatants (lanes 10–18) were analyzed by immunoblot, with the membranes probed with antibodies directed against either Helios (top) or Ikaros (bottom). The two isoforms predominantly expressed in VL3-3M2 cells are indicated to the left of the bottom panel. The amounts of anti-Ikaros or anti-Helios IgGs used in the immunoprecipitations were as follows: 1.5 μg (lanes 2,6,11,,15), 4.5 μg (lanes 3,7,12,16), 15 μg (lanes 4,8,13,,17), and 45 μg (lanes 5,9,14,,18).
Figure 9
The Ikaros–Helios complexes appear to be relatively homogeneous when analyzed by gel-filtration chromatography. Proteins in Superose 6 (Pharmacia) gel-filtration column fractions were separated by 10% SDS-PAGE and analyzed by Western blot analysis involving probing of the blot sequentially with anti-Helios serum (top) followed by anti-Ikaros serum (bottom). Four micrograms of RLm11 nuclear extracts was also analyzed (lane 1). The fractions where standard molecular mass markers migrate in Superose 6 (Pharmacia) gel-filtration chromatography are indicated by arrows on the top with their molecular masses. Isoforms I, III, V and VI are indicated.
Figure 10
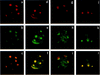
Distribution of Helios and Ikaros proteins within the nuclei of T and B lymphocytes. Confocal images are shown of single optical sections through the nucleus of representative individual concavalin-A stimulated lymph node T cells (a,b,c,g,h,i), activated VL3-3M2 T cells (d,e,f), and B3 pre-B cells (j,k,l). The cells were labeled simultaneously with a probe for gamma satellite sequences (red) and specific antisera for Helios (a–f) or Ikaros (g–l) (shown in green). The red and green components of costained nuclei (bottom, c,f,i,l) are shown separately in the top (gamma satellite-red a,d,g,j) and middle (p70-green b,e, Ikaros-green h,k) rows.
Similar articles
- Targeting of Ikaros to pericentromeric heterochromatin by direct DNA binding.
Cobb BS, Morales-Alcelay S, Kleiger G, Brown KE, Fisher AG, Smale ST. Cobb BS, et al. Genes Dev. 2000 Sep 1;14(17):2146-60. doi: 10.1101/gad.816400. Genes Dev. 2000. PMID: 10970879 Free PMC article. - Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin.
Brown KE, Guest SS, Smale ST, Hahm K, Merkenschlager M, Fisher AG. Brown KE, et al. Cell. 1997 Dec 12;91(6):845-54. doi: 10.1016/s0092-8674(00)80472-9. Cell. 1997. PMID: 9413993 - Helios, a novel dimerization partner of Ikaros expressed in the earliest hematopoietic progenitors.
Kelley CM, Ikeda T, Koipally J, Avitahl N, Wu L, Georgopoulos K, Morgan BA. Kelley CM, et al. Curr Biol. 1998 Apr 23;8(9):508-15. doi: 10.1016/s0960-9822(98)70202-7. Curr Biol. 1998. PMID: 9560339 - Ikaros, Aiolos and Helios: transcription regulators and lymphoid malignancies.
Rebollo A, Schmitt C. Rebollo A, et al. Immunol Cell Biol. 2003 Jun;81(3):171-5. doi: 10.1046/j.1440-1711.2003.01159.x. Immunol Cell Biol. 2003. PMID: 12752680 Review. - Aiolos and Ikaros: regulators of lymphocyte development, homeostasis and lymphoproliferation.
Schmitt C, Tonnelle C, Dalloul A, Chabannon C, Debré P, Rebollo A. Schmitt C, et al. Apoptosis. 2002 Jun;7(3):277-84. doi: 10.1023/a:1015372322419. Apoptosis. 2002. PMID: 11997672 Review.
Cited by
- Germline IKAROS dimerization haploinsufficiency causes hematologic cytopenias and malignancies.
Kuehn HS, Niemela JE, Stoddard J, Mannurita SC, Shahin T, Goel S, Hintermeyer M, Heredia RJ, Garofalo M, Lucas L, Singh S, Tondo A, Jacobs Z, Gahl WA, Latour S, Verbsky J, Routes J, Cunningham-Rundles C, Boztug K, Gambineri E, Fleisher TA, Chandrakasan S, Rosenzweig SD. Kuehn HS, et al. Blood. 2021 Jan 21;137(3):349-363. doi: 10.1182/blood.2020007292. Blood. 2021. PMID: 32845957 Free PMC article. - Expression of Helios in gastric tumor cells predicts better survival in gastric cancer patients.
Chen WM, Wu CS, Liu JL, Yeh CM, Tseng L, Huang HC, Chang PJ, Wu SF. Chen WM, et al. J Cancer Res Clin Oncol. 2016 Nov;142(11):2375-82. doi: 10.1007/s00432-016-2223-3. Epub 2016 Aug 30. J Cancer Res Clin Oncol. 2016. PMID: 27576507 - Targeting of Ikaros to pericentromeric heterochromatin by direct DNA binding.
Cobb BS, Morales-Alcelay S, Kleiger G, Brown KE, Fisher AG, Smale ST. Cobb BS, et al. Genes Dev. 2000 Sep 1;14(17):2146-60. doi: 10.1101/gad.816400. Genes Dev. 2000. PMID: 10970879 Free PMC article. - Helios-controller of Treg stability and function.
Chougnet C, Hildeman D. Chougnet C, et al. Transl Cancer Res. 2016 Aug;5(Suppl 2):S338-S341. doi: 10.21037/tcr.2016.07.37. Transl Cancer Res. 2016. PMID: 30656143 Free PMC article. No abstract available. - The combined expression patterns of Ikaros isoforms characterize different hematological tumor subtypes.
Orozco CA, Acevedo A, Cortina L, Cuellar GE, Duarte M, Martín L, Mesa NM, Muñoz J, Portilla CA, Quijano SM, Quintero G, Rodriguez M, Saavedra CE, Groot H, Torres MM, López-Segura V. Orozco CA, et al. PLoS One. 2013 Dec 6;8(12):e82411. doi: 10.1371/journal.pone.0082411. eCollection 2013. PLoS One. 2013. PMID: 24324784 Free PMC article.
References
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. New York, NY: John Wiley; 1989.
- Babichuk CK, Duggan BL, Bleackley RC. In vivo regulation of murine granzyme B gene transcription in activated primary T cells. J Biol Chem. 1996;271:16485–16493. - PubMed
- Brown KE, Guest SS, Smale ST, Hahm K, Merkenschlager M, Fisher AG. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91:845–854. - PubMed
- Clevers HC, Grosschedl R. Transcriptional control of lymphoid development: Lessons from gene targeting. Immunol Today. 1996;17:336–343. - PubMed
- Clevers HC, Oosterwegel MA, Georgopoulos K. Transcription factors in early T-cell development. Immunol Today. 1993;14:591–596. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM-08042/GM/NIGMS NIH HHS/United States
- T32 GM008042/GM/NIGMS NIH HHS/United States
- GM-07104/GM/NIGMS NIH HHS/United States
- T32 GM007104/GM/NIGMS NIH HHS/United States
- R01 DK043726/DK/NIDDK NIH HHS/United States
- CA442551/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases