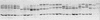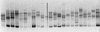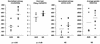Interferon resistance of hepatitis C virus genotype 1b: relationship to nonstructural 5A gene quasispecies mutations - PubMed (original) (raw)
Interferon resistance of hepatitis C virus genotype 1b: relationship to nonstructural 5A gene quasispecies mutations
J M Pawlotsky et al. J Virol. 1998 Apr.
Abstract
A 40-amino-acid sequence located in the nonstructural 5A (NS5A) protein of hepatitis C virus genotype 1b (HCV-1b) was recently suggested to be the interferon sensitivity-determining region (ISDR), because HCV-1b strains with an ISDR amino acid sequence identical to that of the prototype strain HCV-J were found to be resistant to alpha interferon (IFN-alpha) whereas strains with amino acid substitutions were found to be sensitive (N. Enomoto, I. Sakuma, Y. Asahina, M. Kurosaki, T. Murakami, C. Yamamoto, N. Izumi, F. Marumo, and C. Sato, J. Clin. Invest. 96:224-230, 1995; N. Enomoto, I. Sakuma, Y. Asahina, M. Kurosaki, T. Murakami, C. Yamamoto, Y. Ogura, N. Izumi, F. Marumo, and C. Sato, N. Engl. J. Med. 334:77-81, 1996). We used single-strand conformation polymorphism (SSCP) analysis, combined with cloning and sequencing strategies, to characterize NS5A quasispecies in HCV-1b-infected patients and determine the relationships between pre- and posttreatment NS5A quasispecies mutations and the IFN-alpha sensitivity of HCV-1b. The serine residues involved in phosphorylation of NS5A protein were highly conserved both in the various patients and in quasispecies in a given patient, suggesting that phosphorylation is important in NS5A protein function. A hot spot for amino acid substitutions was found at positions 2217 to 2218; it could be the result of either strong selection pressure or tolerance to these amino acid replacements. The proportion of synonymous mutations was significantly higher than the proportion of nonsynonymous mutations, suggesting that genetic variability in the region studied was the result of high mutation rates and viral replication kinetics rather than of positive selection. Sustained HCV RNA clearance was associated with low viral load and low nucleotide sequence entropy, suggesting (i) that the replication kinetics when treatment is started plays a critical role in HCV-1b sensitivity to IFN-alpha and (ii) that HCV-1b resistance to IFN-alpha could be conferred by numerous and/or related mutations that could be patient specific and located at different positions throughout the viral genome and could allow escape variants to be selected by IFN-alpha-stimulated immune responses. No NS5A sequence appeared to be intrinsically resistant or sensitive to IFN-alpha, but the HCV-J sequence was significantly more frequent in nonresponder quasispecies than in sustained virological responder quasispecies, suggesting that the balance between NS5A quasispecies sequences in infected patients could have a subtle regulatory influence on HCV replication.
Figures
FIG. 1
Clonal frequency analysis by SSCP. A 185-bp fragment in the NS5A gene of HCV was amplified by PCR, and the PCR product was cloned into the pTAg vector. Thirty clones were isolated, clonal DNA was amplified by PCR, and the PCR products were analyzed by SSCP. Each clone theoretically gives two bands in SSCP, corresponding to the two strands of double-stranded PCR products, which migrate to positions depending strictly on their nucleotide sequence. At the low temperature used for migration (3°C), minor conformations of DNA strands can generate weak additional bands which also migrate to specific positions. A first SSCP round was performed in random order. The second SSCP round, in which the clones are in the order of frequency deduced from the first analysis, is shown. Each lane corresponds to a different NS5A clone. After clonal frequency analysis, one to three (when available) clones per SSCP pattern were sequenced.
FIG. 2
SSCP analysis of NS5A PCR products obtained from pretreatment samples from HCV-1b-infected patients. One patient’s sample is analyzed per lane. The SSCP patterns were different from one patient to the next and were characterized by the presence of two to eight bands (mean, 3.5 ± 1.5) migrating to different positions in the gel. These patterns suggested that the patients harbored mixes of different sequences in the region of interest, i.e., that this region had a quasispecies distribution.
FIG. 3
Alignment of nucleotide sequences of independent HCV NS5A clones from two patients, characteristic of a quasispecies distribution in the region of interest. Relative frequencies determined by SSCP are shown on the right (the nucleotide sequence between codons 2195 and 2240 of the HCV-1b sequence is shown).
FIG. 4
Amino acid composition (amino acid positions 2195 to 2240) of NS5A protein of quasispecies from 13 patients. The alignment of amino acid sequences was deduced from the nucleotide sequences of 30 to 48 independent clones obtained before IFN-α treatment in 13 patients and 30 to 48 independent clones obtained 6 months after IFN-α withdrawal in 4 of them (patients 10 to 13). Relative frequencies of the variants are shown on the right, as determined by clonal frequency analysis by means of SSCP. Patients 1 to 5 were sustained virological responders to 3 MU of IFN-α three times a week for 6 months; i.e., they cleared HCV during treatment and were still nonviremic 6 months later. Patients 6 to 13 were nonresponders. The HCV-J sequence, reported in 1990 by Kato et al. (33), is given for comparison at the top, and the variants with a sequence identical to HCV-J are identified by an asterisk. The three serine residues mandatory for hyperphosphorylation of the product of the NS5A gene (68) are underlined. Amino acid positions 2217 to 2218, at which nonsynonymous mutations were frequently found, are identified by +.
FIG. 4
Amino acid composition (amino acid positions 2195 to 2240) of NS5A protein of quasispecies from 13 patients. The alignment of amino acid sequences was deduced from the nucleotide sequences of 30 to 48 independent clones obtained before IFN-α treatment in 13 patients and 30 to 48 independent clones obtained 6 months after IFN-α withdrawal in 4 of them (patients 10 to 13). Relative frequencies of the variants are shown on the right, as determined by clonal frequency analysis by means of SSCP. Patients 1 to 5 were sustained virological responders to 3 MU of IFN-α three times a week for 6 months; i.e., they cleared HCV during treatment and were still nonviremic 6 months later. Patients 6 to 13 were nonresponders. The HCV-J sequence, reported in 1990 by Kato et al. (33), is given for comparison at the top, and the variants with a sequence identical to HCV-J are identified by an asterisk. The three serine residues mandatory for hyperphosphorylation of the product of the NS5A gene (68) are underlined. Amino acid positions 2217 to 2218, at which nonsynonymous mutations were frequently found, are identified by +.
FIG. 5
Nucleotide sequence entropy, amino acid sequence entropy, average genetic distances within the quasispecies, and viral load in the five patients with a sustained virological response (SVR) to IFN-α therapy and in the eight nonresponders (NR). NS, not significantly different.
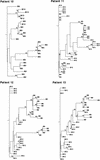
FIG. 6
Phylogenetic trees of the NS5A region within the four subjects studied before IFN-α treatment and 12 months later. The phylogenetic reconstructions shown are neighbor-joining trees with bootstrap proportions of greater than 50 of 100 bootstrap replicates shown at appropriate branch points. M0 indicates a variant isolated before treatment, and M12 indicates a variant isolated 12 months later.
Similar articles
- Quasispecies evolution in NS5A region of hepatitis C virus genotype 1b during interferon or combined interferon-ribavirin therapy.
Veillon P, Payan C, Le Guillou-Guillemette H, Gaudy C, Lunel F. Veillon P, et al. World J Gastroenterol. 2007 Feb 28;13(8):1195-203. doi: 10.3748/wjg.v13.i8.1195. World J Gastroenterol. 2007. PMID: 17451199 Free PMC article. - Evidence for sequence selection within the non-structural 5A gene of hepatitis C virus type 1b during unsuccessful treatment with interferon-alpha.
Gerotto M, Dal Pero F, Sullivan DG, Chemello L, Cavalletto L, Polyak SJ, Pontisso P, Gretch DR, Alberti A. Gerotto M, et al. J Viral Hepat. 1999 Sep;6(5):367-72. doi: 10.1046/j.1365-2893.1999.00166.x. J Viral Hepat. 1999. PMID: 10607252 - Mutations in ISDR of NS5A gene influence interferon efficacy in Chinese patients with chronic hepatitis C virus genotype 1b infection.
Shen C, Hu T, Shen L, Gao L, Xie W, Zhang J. Shen C, et al. J Gastroenterol Hepatol. 2007 Nov;22(11):1898-903. doi: 10.1111/j.1440-1746.2006.04566.x. J Gastroenterol Hepatol. 2007. PMID: 17914967 - The non-structural 5A protein of hepatitis C virus.
Pawlotsky JM, Germanidis G. Pawlotsky JM, et al. J Viral Hepat. 1999 Sep;6(5):343-56. doi: 10.1046/j.1365-2893.1999.00185.x. J Viral Hepat. 1999. PMID: 10607250 Review. - Repression of the PKR protein kinase by the hepatitis C virus NS5A protein: a potential mechanism of interferon resistance.
Gale MJ Jr, Korth MJ, Katze MG. Gale MJ Jr, et al. Clin Diagn Virol. 1998 Jul 15;10(2-3):157-62. doi: 10.1016/s0928-0197(98)00034-8. Clin Diagn Virol. 1998. PMID: 9741641 Review.
Cited by
- Direct-Acting Antivirals Reduce the De Novo Development of Esophageal Varices in Patients with Hepatitis C Virus Related Liver Cirrhosis.
Hsieh YY, Chen WM, Chang KC, Chang TS, Hung CH, Yang YH, Tung SY, Wei KL, Shen CH, Wu CS, Ding YJ, Hu JH, Huang YT, Lin MH, Lu CK, Lin YH, Lin MS. Hsieh YY, et al. Viruses. 2023 Jan 16;15(1):252. doi: 10.3390/v15010252. Viruses. 2023. PMID: 36680293 Free PMC article. - The Relationship Between HCV-NS5A Gene Mutations and Resistance to Combination Therapy in Patients with HCV- Genotype 1-B.
Lashgarian HE, Valibeik A, Marzban A, Karkhane M, Shahzamani K. Lashgarian HE, et al. Rep Biochem Mol Biol. 2021 Jul;10(2):233-242. doi: 10.52547/rbmb.10.2.233. Rep Biochem Mol Biol. 2021. PMID: 34604413 Free PMC article. - Induction of Combination Therapy for the Management of Hepatitis C: An Observational Study.
Khan SZ, Talha MU, Iftikhar B, Noor A, Laique T, Latif A, Malik J. Khan SZ, et al. Cureus. 2020 Sep 5;12(9):e10259. doi: 10.7759/cureus.10259. Cureus. 2020. PMID: 33042697 Free PMC article. - Analysis of mutations in the core and NS5A genes of hepatitis C virus in non-responder and relapser patients after treatment with Peg-IFN-α and ribavirin.
Kumthip K, Chusri P, Pantip C, Thongsawat S, O'Brien A, Maneekarn N. Kumthip K, et al. Virusdisease. 2016 Mar;27(1):55-62. doi: 10.1007/s13337-015-0300-x. Epub 2016 Jan 18. Virusdisease. 2016. PMID: 26925445 Free PMC article. - Nonstructural 5A Protein of Hepatitis C Virus Regulates Soluble Resistance-Related Calcium-Binding Protein Activity for Viral Propagation.
Tran GV, Luong TT, Park EM, Kim JW, Choi JW, Park C, Lim YS, Hwang SB. Tran GV, et al. J Virol. 2015 Dec 30;90(6):2794-805. doi: 10.1128/JVI.02493-15. J Virol. 2015. PMID: 26719254 Free PMC article.
References
- Alter H J. To C or not to C: these are the questions. Blood. 1995;85:1681–1695. - PubMed
- Baron S, Tyring S K, Fleischmann W R, Coppenhaver D H, Niesel D W, Klimpel G R, Stanton G J, Hughes T K. The interferons. Mechanisms of action and clinical applications. JAMA. 1991;266:1375–1383. - PubMed
- Chayama K, Tsubota A, Kobayashi M, Okamoto K, Hashimoto M, Miyano Y, Koike H, Kobayashi M, Koida I, Arase Y, Saitoh S, Suzuki Y, Murashima N, Ikeda K, Kumada H. Pretreatment virus load and multiple amino acid substitutions in the interferon sensitivity-determining region predict the outcome of interferon treatment in patients with chronic genotype 1b hepatitis C virus infection. Hepatology. 1997;25:745–749. - PubMed
- Chemello L, Cavalletto L, Casarin C, Bonetti P, Bernardinello E, Pontisso P, Donada C, Belussi F, Martinelli S, Alberti A the TriVeneto Viral Hepatitis Group. Persistent hepatitis C viremia predicts late relapse after sustained response to interferon-α in chronic hepatitis C. Ann Intern Med. 1996;124:1058–1060. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases