Coupled transcriptional and translational control of cyclin-dependent kinase inhibitor p18INK4c expression during myogenesis - PubMed (original) (raw)
Coupled transcriptional and translational control of cyclin-dependent kinase inhibitor p18INK4c expression during myogenesis
D E Phelps et al. Mol Cell Biol. 1998 Apr.
Abstract
Terminal differentiation of many cell types involves permanent withdrawal from the cell division cycle. The p18INK4c protein, a member of the p16/INK4 cyclin-dependent kinase (CDK) inhibitor family, is induced more than 50-fold during myogenic differentiation of mouse C2C12 myoblasts to become the predominant CDK inhibitor complexed with CDK4 and CDK6 in terminally differentiated myotubes. We have found that the p18INK4c gene expresses two mRNA transcripts--a 2.4-kb transcript, p18(L), and a 1.2-kb transcript, p18(S). In proliferating C2C12 myoblasts, only the larger p18(L) transcript is expressed from an upstream promoter. As C2C12 cells are induced to differentiate into permanently arrested myotubes, the abundance of the p18(L) transcript decreases. The smaller p18(S) transcript expressed from a downstream promoter becomes detectable by 12 h postinduction and is the predominant transcript expressed in terminally differentiated myotubes. Both transcripts contain coding exons 2 and 3, but p18(L) uniquely contains an additional noncoding 1.2-kb exon, exon 1, corresponding exclusively to the 5' untranslated region (5' UTR). The expression pattern of the shorter p18(S) transcript, but not that of the longer p18(L) transcript, correlates with terminal differentiation of muscle, lung, liver, thymus, and eye lens cells during mouse embryo development. The presence of the long 5' UTR in exon 1 attenuated the translation of p18(L) transcript, while its absence from the shorter p18(S) transcript resulted in significantly more efficient translation of the p18 protein. Our results demonstrate that during terminal muscle cell differentiation, induction of the p18 protein is regulated by promoter switching coupled with translational control.
Figures
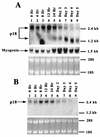
FIG. 1
Differential expression of p18 mRNA during C2C12 cell differentiation. (A) Total RNA was prepared from proliferating C2C12 cells cultured in growth medium (lane 1) or in differentiation medium for the time indicated above each lane (lanes 2 to 9). Ten micrograms of each RNA sample was resolved on a 1% agarose–formaldehyde–MOPS gel, transferred to a nitrocellulose filter, and hybridized with a probe derived from the coding region of mouse p18 cDNA (top). The same blot was stripped and rehybridized with a probe derived from the mouse myogenin gene to monitor the progression of myogenesis (middle). Approximately equal amounts of RNA were loaded, as determined by ethidium bromide staining (bottom). (B) Expression of p18 exon 1 during C2C12 cell differentiation. Ten micrograms of total RNA prepared from C2C12 cells cultured in growth medium (lane 1) or in differentiation medium for the time indicated above each lane (lanes 2 to 9) were resolved on a 1% agarose–formaldehyde–MOPS gel, transferred to a nitrocellulose filter, and hybridized with a probe derived from exon 1 of NIH 3T3 cDNA clone T9 (top). Approximately equal amounts of RNA were loaded, as determined by ethidium bromide staining (bottom).
FIG. 2
Genomic and cDNA structures of the mouse p18 gene. (A) Comparison of p18 cDNA 5′ UTRs with the p18 genomic sequence. Nucleotide sequences of the 5′ UTR of a mouse p18 genomic fragment and the p18 cDNAs isolated from proliferating NIH 3T3 cells [cDNA T9, p18(L)] and mature skeletal muscle cells [cDNA M1, p18(S)] are aligned. The splice donor and acceptor sites are boldfaced and underlined. The ATG translation initiation codon for p18 is boldfaced, and other ATG codons encoding short ORFs present in the 5′ UTR (uORFs) of cDNA T9 are italicized and underlined. Two single-nucleotide polymorphisms between the genomic and skeletal muscle sequences are indicated with asterisks. The antisense oligonucleotide primer, L, used for the primer extension experiments is underlined on the coding strands. (B) Schematic representation of the mouse p18 genomic and cDNA structures. The 5′ UTRs of the p18(L) and p18(S) transcripts are indicated by thin- and thick-striped boxes, respectively, the 3′UTR is indicated by grey boxes, introns are indicated by white boxes, and coding regions are indicated by black boxes. Splicing events are indicated by bridged lines.
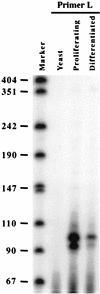
FIG. 3
Primer extension of the p18(L) transcript. Total RNA was prepared from C2C12 cells cultured in growth medium (Proliferating) and C2C12 cells cultured in differentiation medium for 4 days (Differentiated). Yeast tRNA was used as a negative control. Thirty micrograms of each RNA sample was hybridized with an antisense primer specific to the p18(L) transcript, primer L, and extended with reverse transcriptase. The extension products were resolved on a 7% urea denaturing gel. The marker lane is pUC18 digested with _Hpa_II and 5′ end labeled. The fragment lengths (in nucleotides) are indicated on the left.
FIG. 4
The mouse p18 gene contains two promoters. DNA fragments containing mouse p18 genomic sequences were subcloned into the pGL2-Basic luciferase reporter plasmid, which lacks eukaryotic promoter and enhancer sequences. The restriction sites used in the construction of these plasmids are indicated. The A nucleotide in the ATG codon has been arbitrarily set to 1. The promoter activity of each construct was determined by measuring the luciferase activity of each construct 24 h after transient transfection into proliferating NIH 3T3 cells. Transfection efficiency was normalized by including a CMV-LacZ expression plasmid. Relative luciferase activity was normalized to β-galactosidase activity, and the normalized luciferase activity of the promoterless pGL2-Basic plasmid was set to 1. The data are averages of three independent experiments. The transcription initiation site of the p18(S) transcript, localized by promoter activity assays between the _Sma_I (nt 854) and _Hin_dIII (nt 355) restriction sites (highlighted with a dashed line), has not been precisely determined. (A) Localization of a second mouse p18 promoter; (B) comparison of two promoter strengths.
FIG. 5
Expression of p18 mRNA in mouse embryos. Sagittal sections from E15.0 embryos were hybridized with the mouse p18 antisense RNA probe corresponding to the coding sequences common to both long and short transcripts (A, C, and E) or the 5′ UTR uniquely present in the long transcript (G, I, and K). Both dark-field (left) and corresponding bright-field (right) photographs are shown. Testis tissue was from 2-month-old mice (K and L). dp, diaphragm; ht, heart; lv, liver; lu, lung; sc, spinal cord; sm, skeletal muscle; ST, seminiferous tubule; ty, thymus; vc, vertebral cartilage.
FIG. 6
Translational regulation of p18 mRNA by the 5′ UTR. (A) Diagram of the relevant regions of the pcDNA3-p18(L) and pcDNA3-p18(S) plasmids. A 1,840-bp DNA fragment corresponding to the p18(L) cDNA isolated from NIH 3T3 cells and an 845-bp DNA fragment corresponding to the p18(S) cDNA isolated from mouse skeletal muscle tissue were inserted into the pcDNA3 expression vector downstream of the heterologous CMV and phage T7 promoters. The 5′ UTRs are indicated by gray boxes, the 3′ UTRs are indicated by white boxes, and the p18 coding sequences are indicated by black boxes. The positions of the translation initiation codons (ATG) and stop codons (TGA) are indicated. (B) In vitro transcription of the two p18 cDNAs. Both pcDNA3-p18(L) and pcDNA3-p18(S) (0.1 pmol of each) were used as templates for in vitro transcription of 5′ capped mRNA using phage T7 RNA polymerase. The RNA products were verified by loading 0.1 (lanes 1 and 4), 0.25 (lanes 2 and 5), or 0.5 (lanes 3 and 6) μl of each reaction mixture onto a 1% agarose–formaldehyde–MOPS gel and quantitated by spectrophotometry (lanes 1 to 6, respectively, contained 180, 450, 900, 160, 400, and 800 ng). A discrete RNA transcript consistent with the size of the appropriate cloned cDNA fragment was produced from each reaction, as indicated to the left of the gel. (C) In vitro translation of the two p18 cDNAs. The in vitro-transcribed p18 mRNAs (0.1 or 0.5 pmol of each) were in vitro translated in the presence of [35S]methionine. Ten microliters of each translation reaction mixture was immunoprecipitated with antiserum against p18 and resolved by SDS-PAGE. (D and E) C2C12 cells were transiently transfected with the indicated p18 expression plasmid or the parental pcDNA3 plasmid. Cells were harvested 24 h posttransfection and divided into two equal fractions for RNA (D) and protein (E) analyses. Ten micrograms of total RNA from the transfected C2C12 cells was resolved on a 1% agarose–formaldehyde–MOPS gel, transferred to a nylon membrane, and hybridized with a probe derived from the coding region of mouse p18 cDNA (top). Approximately equal amounts of RNA were loaded, as determined by stripping and reprobing of the same blot with a human β-actin probe (bottom). (E) Fifty micrograms of total protein lysate from the transfected C2C12 cells was resolved by SDS-PAGE, transferred to a nitrocellulose filter, and blotted with an antibody specific for the p18 (top) or CDK2 (bottom) protein to verify equal protein loading.
References
- Chin Y E, Kitagawa M, Su W-C S, You Z-H, Iwamoto Y, Fu X-Y. Cell growth arrest and induction of cyclin-dependent kinase inhibitor p21WAF1/CIP1 mediated by STAT1. Science. 1996;272:719–722. - PubMed
- Cox K H, DeLeon D V, Angerer L A, Angerer R C. Detection of mRNA in sea urchin embryos by in situ hybridization using asymmetric RNA probes. Dev Biol. 1984;101:485–502. - PubMed
- Curtis D, Lehmann R, Zamore P D. Translational regulation in development. Cell. 1995;81:171–178. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials